Modification of Pseudomonas aeruginosa Pa5196 type IV Pilins at multiple sites with D-Araf by a novel GT-C family Arabinosyltransferase, TfpW
- PMID: 18805982
- PMCID: PMC2576659
- DOI: 10.1128/JB.01075-08
Modification of Pseudomonas aeruginosa Pa5196 type IV Pilins at multiple sites with D-Araf by a novel GT-C family Arabinosyltransferase, TfpW
Abstract
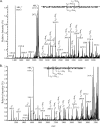
Pseudomonas aeruginosa Pa5196 produces type IV pilins modified with unusual alpha1,5-linked d-arabinofuranose (alpha1,5-D-Araf) glycans, identical to those in the lipoarabinomannan and arabinogalactan cell wall polymers from Mycobacterium spp. In this work, we identify a second strain of P. aeruginosa, PA7, capable of expressing arabinosylated pilins and use a combination of site-directed mutagenesis, electrospray ionization mass spectrometry (MS), and electron transfer dissociation MS to identify the exact sites and extent of pilin modification in strain Pa5196. Unlike previously characterized type IV pilins that are glycosylated at a single position, those from strain Pa5196 were modified at multiple sites, with modifications of alphabeta-loop residues Thr64 and Thr66 being important for normal pilus assembly. Trisaccharides of alpha1,5-D-Araf were the principal modifications at Thr64 and Thr66, with additional mono- and disaccharides identified on Ser residues within the antiparallel beta sheet region of the pilin. TfpW was hypothesized to encode the pilin glycosyltransferase based on its genetic linkage to the pilin, weak similarity to membrane-bound GT-C family glycosyltransferases (which include the Mycobacterium arabinosyltransferases EmbA/B/C), and the presence of characteristic motifs. Loss of TfpW or mutation of key residues within the signature GT-C glycosyltransferase motif completely abrogated pilin glycosylation, confirming its involvement in this process. A Pa5196 pilA mutant complemented with other Pseudomonas pilins containing potential sites of modification expressed nonglycosylated pilins, showing that TfpW's pilin substrate specificity is restricted. TfpW is the prototype of a new type IV pilin posttranslational modification system and the first reported gram-negative member of the GT-C glycosyltransferase family.
Figures








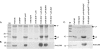

References
-
- Aas, F. E., W. Egge-Jacobsen, H. C. Winther-Larsen, C. Lovold, P. G. Hitchen, A. Dell, and M. Koomey. 2006. Neisseria gonorrhoeae type IV pili undergo multisite, hierarchical modifications with phosphoethanolamine and phosphocholine requiring an enzyme structurally related to lipopolysaccharide phosphoethanolamine transferases. J. Biol. Chem. 28127712-27723. - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

