SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli
- PMID: 18805985
- PMCID: PMC2576670
- DOI: 10.1128/JB.00945-08
SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli
Abstract
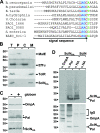
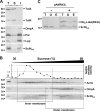
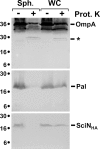
Enteroaggregative Escherichia coli (EAEC) is a pathogen implicated in several infant diarrhea or diarrheal outbreaks in areas of endemicity. Although multiple genes involved in EAEC pathogenesis have been identified, the overall mechanism of virulence is not well understood. Recently, a novel secretion system, called type VI secretion (T6S) system (T6SS), has been identified in EAEC and most animal or plant gram-negative pathogens. T6SSs are multicomponent cell envelope machines responsible for the secretion of at least two putative substrates, Hcp and VgrG. In EAEC, two copies of T6S gene clusters, called sci-1 and sci-2, are present on the pheU pathogenicity island. In this study, we focused our work on the sci-1 gene cluster. The Sci-1 apparatus is probably composed of all, or a subset of, the 21 gene products encoded on the cluster. Among these subunits, some are shared by all T6SSs identified to date, including a ClpV-type AAA(+) ATPase (SciG) and an IcmF (SciS) and an IcmH (SciP) homologue, as well as a putative lipoprotein (SciN). In this study, we demonstrate that sciN is a critical gene necessary for T6S-dependent secretion of the Hcp-like SciD protein and for biofilm formation. We further show that SciN is a lipoprotein, as shown by the inhibition of its processing by globomycin and in vivo labeling with [(3)H]palmitic acid. SciN is tethered to the outer membrane and exposed in the periplasm. Sequestration of SciN at the inner membrane by targeting the +2 residue responsible for lipoprotein localization (Gly2Asp) fails to complement an sciN mutant for SciD secretion and biofilm formation. Together, these results support a model in which SciN is an outer membrane lipoprotein exposed in the periplasm and essential for the Sci-1 apparatus function.
Figures


References
-
- Anyanful, A., J. M. Dolan-Livengood, T. Lewis, S. Sheth, M. N. Dezalia, M. A. Sherman, L. V. Kalman, G. M. Benian, and D. Kalman. 2005. Paralysis and killing of Caenorhabditis elegans by enteropathogenic Escherichia coli requires the bacterial tryptophanase gene. Mol. Microbiol. 57988-1007. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

