TAp73 knockout shows genomic instability with infertility and tumor suppressor functions
- PMID: 18805989
- PMCID: PMC2559903
- DOI: 10.1101/gad.1695308
TAp73 knockout shows genomic instability with infertility and tumor suppressor functions
Abstract
The Trp53 gene family member Trp73 encodes two major groups of protein isoforms, TAp73 and DeltaNp73, with opposing pro- and anti-apoptotic functions; consequently, their relative ratio regulates cell fate. However, the precise roles of p73 isoforms in cellular events such as tumor initiation, embryonic development, and cell death remain unclear. To determine which aspects of p73 function are attributable to the TAp73 isoforms, we generated and characterized mice in which exons encoding the TAp73 isoforms were specifically deleted to create a TAp73-deficient (TAp73(-/-)) mouse. Here we show that mice specifically lacking in TAp73 isoforms develop a phenotype intermediate between the phenotypes of Trp73(-/-) and Trp53(-/-) mice with respect to incidence of spontaneous and carcinogen-induced tumors, infertility, and aging, as well as hippocampal dysgenesis. In addition, cells from TAp73(-/-) mice exhibit genomic instability associated with enhanced aneuploidy, which may account for the increased incidence of spontaneous tumors observed in these mutants. Hence, TAp73 isoforms exert tumor-suppressive functions and indicate an emerging role for Trp73 in the maintenance of genomic stability.
Figures
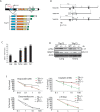
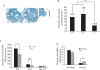
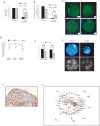



Comment in
-
The jury is in: p73 is a tumor suppressor after all.Genes Dev. 2008 Oct 1;22(19):2591-5. doi: 10.1101/gad.1727408. Genes Dev. 2008. PMID: 18832062 Review.
References
-
- Acton B.M., Jurisicova A., Jurisica I., Casper R.F. Alterations in mitochondrial membrane potential during preimplantation stages of mouse and human embryo development. Mol. Hum. Reprod. 2004;10:23–32. - PubMed
-
- Baird D.T., Collins J., Egozcue J., Evers L.H., Gianaroli L., Leridon H., Sunde A., Templeton A., Van Steirteghem A., Cohen J., et al. Fertility and ageing. Hum. Reprod. Update. 2005;11:261–276. - PubMed
-
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics. 2003;19:185–193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
