ZNF198 stabilizes the LSD1-CoREST-HDAC1 complex on chromatin through its MYM-type zinc fingers
- PMID: 18806873
- PMCID: PMC2532748
- DOI: 10.1371/journal.pone.0003255
ZNF198 stabilizes the LSD1-CoREST-HDAC1 complex on chromatin through its MYM-type zinc fingers
Abstract
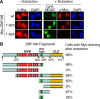
Histone modifications in chromatin regulate gene expression. A transcriptional co-repressor complex containing LSD1-CoREST-HDAC1 (termed LCH hereafter for simplicity) represses transcription by coordinately removing histone modifications associated with transcriptional activation. RE1-silencing transcription factor (REST) recruits LCH to the promoters of neuron-specific genes, thereby silencing their transcription in non-neuronal tissues. ZNF198 is a member of a family of MYM-type zinc finger proteins that associate with LCH. Here, we show that ZNF198-like proteins are required for the repression of E-cadherin (a gene known to be repressed by LSD1), but not REST-responsive genes. ZNF198 binds preferentially to the intact LCH ternary complex, but not its individual subunits. ZNF198- and REST-binding to the LCH complex are mutually exclusive. ZNF198 associates with chromatin independently of LCH. Furthermore, modification of HDAC1 by small ubiquitin-like modifier (SUMO) in vitro weakens its interaction with CoREST whereas sumoylation of HDAC1 stimulates its binding to ZNF198. Finally, we mapped the LCH- and HDAC1-SUMO-binding domains of ZNF198 to tandem repeats of MYM-type zinc fingers. Therefore, our results suggest that ZNF198, through its multiple protein-protein interaction interfaces, helps to maintain the intact LCH complex on specific, non-REST-responsive promoters and may also prevent SUMO-dependent dissociation of HDAC1.
Conflict of interest statement
Figures






References
-
- Luger K. Structure and dynamic behavior of nucleosomes. Curr Opin Genet Dev. 2003;13:127–135. - PubMed
-
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol. 2002;319:1097–1113. - PubMed
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. - PubMed
-
- Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr Opin Struct Biol. 2005;15:188–196. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

