Evidence for positive selection acting on microcystin synthetase adenylation domains in three cyanobacterial genera
- PMID: 18808704
- PMCID: PMC2564945
- DOI: 10.1186/1471-2148-8-256
Evidence for positive selection acting on microcystin synthetase adenylation domains in three cyanobacterial genera
Abstract
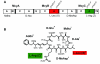
Background: Cyanobacteria produce a wealth of secondary metabolites, including the group of small cyclic heptapeptide hepatotoxins that constitutes the microcystin family. The enzyme complex that directs the biosynthesis of microcystin is encoded in a single large gene cluster (mcy). mcy genes have a widespread distribution among cyanobacteria and are likely to have an ancient origin. The notable diversity within some of the Mcy modules is generated through various recombination events including horizontal gene transfer.
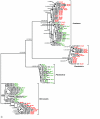
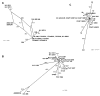
Results: A comparative analysis of the adenylation domains from the first module of McyB (McyB1) and McyC in the microcystin synthetase complex was performed on a large number of microcystin-producing strains from the Anabaena, Microcystis and Planktothrix genera. We found no decisive evidence for recombination between strains from different genera. However, we detected frequent recombination events in the mcyB and mcyC genes between strains within the same genus. Frequent interdomain recombination events were also observed between mcyB and mcyC sequences in Anabaena and Microcystis. Recombination and mutation rate ratios suggest that the diversification of mcyB and mcyC genes is driven by recombination events as well as point mutations in all three genera. Sequence analysis suggests that generally the adenylation domains of the first domain of McyB and McyC are under purifying selection. However, we found clear evidence for positive selection acting on a number of amino acid residues within these adenylation domains. These include residues important for active site selectivity of the adenylation domain, strongly suggesting selection for novel microcystin variants.
Conclusion: We provide the first clear evidence for positive selection acting on amino acid residues involved directly in the recognition and activation of amino acids incorporated into microcystin, indicating that the microcystin complement of a given strain may influence the ability of a particular strain to interact with its environment.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

