MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings
- PMID: 18809224
- PMCID: PMC5303623
- DOI: 10.1016/j.tig.2008.08.007
MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings
Abstract
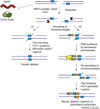
DNA double-strand breaks are normal consequences of cell division and differentiation and must be repaired faithfully to maintain genome stability. Two mechanistically distinct pathways are known to efficiently repair double-strand breaks: homologous recombination and Ku-dependent non-homologous end joining. Recently, a third, less characterized repair mechanism, named microhomology-mediated end joining (MMEJ), has received increasing attention. MMEJ repairs DNA breaks via the use of substantial microhomology and always results in deletions. Furthermore, it probably contributes to oncogenic chromosome rearrangements and genetic variation in humans. Here, we summarize the genetic attributes of MMEJ from several model systems and discuss the relationship between MMEJ and 'alternative end joining'. We propose a mechanistic model for MMEJ and highlight important questions for future research.
Figures

References
-
- Keeney S, Neale MJ. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem. Soc. Trans. 2006;34:523–525. - PubMed
-
- Jung D, et al. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 2006;24:541–570. - PubMed
-
- Shrivastav M, et al. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. - PubMed
-
- Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell. 2007;131:223–225. - PubMed
-
- Chen C, et al. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol. Cell. 1998;2:9–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

