Histone modifications, but not nucleosomal positioning, correlate with major histocompatibility complex class I promoter activity in different tissues in vivo
- PMID: 18809568
- PMCID: PMC2593446
- DOI: 10.1128/MCB.00889-08
Histone modifications, but not nucleosomal positioning, correlate with major histocompatibility complex class I promoter activity in different tissues in vivo
Abstract
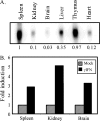
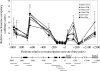
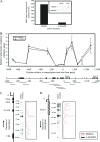
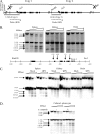
To examine the role of chromatin in transcriptional regulation of the major histocompatibility complex (MHC) class I gene, we determined nucleosome occupancy and positioning, histone modifications, and H2A.Z occupancy across its regulatory region in murine tissues that have widely different expression levels. Surprisingly, nucleosome occupancy and positioning were indistinguishable between the spleen, kidney, and brain. In all three tissues, the 200 bp upstream of the transcription start site had low nucleosome occupancy. In contrast, nuclease hypersensitivity, histone modifications, and H2A.Z occupancy showed tissue-specific differences. Thus, tissue-specific differences in MHC class I transcription correlate with histone modifications and not nucleosomal organization. Further, activation of class I transcription by gamma interferon or its inhibition by alpha-amanitin did not alter nucleosome occupancy, positioning, nuclease hypersensitivity, histone modifications, or H2A.Z occupancy in any of the tissues examined. Thus, chromatin remodeling was not required to dynamically modulate transcriptional levels. These findings suggest that the MHC class I promoter remains poised and accessible to rapidly respond to infection and environmental cues.
Figures


References
-
- Albert, I., T. N. Mavrich, L. P. Tomsho, J. Qi, S. J. Zanton, S. C. Schuster, and B. F. Pugh. 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446572-576. - PubMed
-
- Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
