PML activates transcription by protecting HIPK2 and p300 from SCFFbx3-mediated degradation
- PMID: 18809579
- PMCID: PMC2593379
- DOI: 10.1128/MCB.00897-08
PML activates transcription by protecting HIPK2 and p300 from SCFFbx3-mediated degradation
Abstract
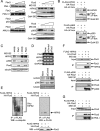
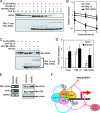
PML, a nuclear protein, interacts with several transcription factors and their coactivators, such as HIPK2 and p300, resulting in the activation of transcription. Although PML is thought to achieve transcription activation by stabilizing the transcription factor complex, little is known about the underlying molecular mechanism. To clarify the role of PML in transcription regulation, we purified the PML complex and identified Fbxo3 (Fbx3), Skp1, and Cullin1 as novel components of this complex. Fbx3 formed SCF(Fbx3) ubiquitin ligase and promoted the degradation of HIPK2 and p300 by the ubiquitin-proteasome pathway. PML inhibited this degradation through a mechanism that unexpectedly did not involve inhibition of the ubiquitination of HIPK2. PML, Fbx3, and HIPK2 synergistically activated p53-induced transcription. Our findings suggest that PML stabilizes the transcription factor complex by protecting HIPK2 and p300 from SCF(Fbx3)-induced degradation until transcription is completed. In contrast, the leukemia-associated fusion PML-RARalpha induced the degradation of HIPK2. We discuss the roles of PML and PML-retinoic acid receptor alpha, as well as those of HIPK2 and p300 ubiquitination, in transcriptional regulation and leukemogenesis.
Figures





References
-
- Avantaggiati, M. L., V. Ogryzko, K. Gardner, A. Giordano, A. S. Levine, and K. Kelly. 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 891175-1184. - PubMed
-
- Bernardi, R., P. P. Scaglioni, S. Bergmann, H. F. Horn, K. H. Vousden, and P. P. Pandolfi. 2004. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat. Cell Biol. 6665-672. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
