Distinct sets of alphabeta TCRs confer similar recognition of tumor antigen NY-ESO-1157-165 by interacting with its central Met/Trp residues
- PMID: 18809922
- PMCID: PMC2567484
- DOI: 10.1073/pnas.0807954105
Distinct sets of alphabeta TCRs confer similar recognition of tumor antigen NY-ESO-1157-165 by interacting with its central Met/Trp residues
Abstract
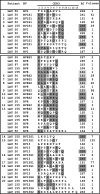
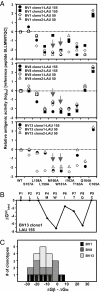
Naturally acquired immune responses against human cancers often include CD8(+) T cells specific for the cancer testis antigen NY-ESO-1. Here, we studied T cell receptor (TCR) primary structure and function of 605 HLA-A*0201/NY-ESO-1(157-165)-specific CD8 T cell clones derived from five melanoma patients. We show that an important proportion of tumor-reactive T cells preferentially use TCR AV3S1/BV8S2 chains, with remarkably conserved CDR3 amino acid motifs and lengths in both chains. All remaining T cell clones belong to two additional sets expressing BV1 or BV13 TCRs, associated with alpha-chains with highly diverse VJ usage, CDR3 amino acid sequence, and length. Yet, all T cell clonotypes recognize tumor antigen with similar functional avidity. Two residues, Met-160 and Trp-161, located in the middle region of the NY-ESO-1(157-165) peptide, are critical for recognition by most of the T cell clonotypes. Collectively, our data show that a large number of alphabeta TCRs, belonging to three distinct sets (AVx/BV1, AV3/BV8, AVx/BV13) bind pMHC with equal antigen sensitivity and recognize the same peptide motif. Finally, this in-depth study of recognition of a self-antigen suggests that in part similar biophysical mechanisms shape TCR repertoires toward foreign and self-antigens.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Zinkernagel RM, Doherty PC. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature. 1974;248:701–702. - PubMed
-
- van der Merwe PA, Davis SJ. Molecular interactions mediating T cell antigen recognition. Annu Rev Immunol. 2003;21:659–684. - PubMed
-
- Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T cell repertoire diversity. Nat Rev Immunol. 2004;4:123–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

