Genetic heterogeneity in pbp genes among clinically isolated group B Streptococci with reduced penicillin susceptibility
- PMID: 18809936
- PMCID: PMC2592870
- DOI: 10.1128/AAC.00596-08
Genetic heterogeneity in pbp genes among clinically isolated group B Streptococci with reduced penicillin susceptibility
Abstract
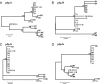
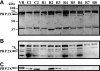
The recent emergence of group B streptococcal isolates exhibiting increased penicillin MICs at the Funabashi Municipal Medical Center and other hospitals in Japan prompted a comparative analysis of the penicillin-binding proteins (PBPs) from those strains with the PBPs from penicillin-susceptible strains comprising four neonatal invasive strains isolated from 1976 to 1988 and two recent isolates. The PBP sequences of the penicillin-susceptible strains were highly conserved, irrespective of their isolation date. Of six strains with reduced susceptibility to penicillin (penicillin MICs, 0.25 to 0.5 mug/ml), strains R1, R2, R5, and R6 shared a unique set of five amino acid substitutions, including V405A adjacent to the (402)SSN(404) motif in PBP 2X and one in PBP 2B. The remaining two strains, R3 and R4, shared several substitutions, including Q557E adjacent to the (552)KSG(554) motif in PBP 2X, in addition to the substitutions in PBP 2B, which are commonly found among penicillin-insusceptible strains. Strains R7 and R8, which had a penicillin MIC of 1 mug/ml, shared a unique set of eight amino acid substitutions (two in PBP 2X; two in PBP 2B, including G613R adjacent to the (614)KTG(616) motif; three in PBP 1A; and one in PBP 2A), and the Q557E substitution in PBP 2X was common to R3 and R4. The binding of Bocillin FL was reduced or not detected in some PBPs, including PBP 2X of penicillin-insusceptible strains, but no significant reduction in the level of pbp2x transcription was found in such strains. The results of phylogenetic comparative analyses imply the absence of epidemic penicillin-insusceptible strains, and several genetic lineages of penicillin-insusceptible strains have been independently emerging through the accumulation of mutations in their pbp genes, especially in pbp2x.
Figures


References
-
- Appelbaum, P. C. 1992. Antimicrobial resistance in Streptococcus pneumoniae: an overview. Clin. Infect. Dis. 15:77-83. - PubMed
-
- Appelbaum, P. C., A. Bhamjee, J. N. Scragg, A. F. Hallett, A. J. Bowen, and R. C. Cooper. 1977. Streptococcus pneumoniae resistant to penicillin and chloramphenicol. Lancet ii:995-997. - PubMed
-
- Biedenbach, D. J., J. M. Stephen, and R. N. Jones. 2003. Antimicrobial susceptibility profile among beta-haemolytic Streptococcus spp. collected in the SENTRY Antimicrobial Surveillance Program—North America, 2001. Diagn. Microbiol. Infect. Dis. 46:291-294. - PubMed
-
- Clinical and Laboratory Standards Institute. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A6, 6th ed. Clinical and Laboratory Standards Institute, Wayne, PA.
-
- Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing. Supplement M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

