The Serine/threonine kinase Stk33 exhibits autophosphorylation and phosphorylates the intermediate filament protein Vimentin
- PMID: 18811945
- PMCID: PMC2567967
- DOI: 10.1186/1471-2091-9-25
The Serine/threonine kinase Stk33 exhibits autophosphorylation and phosphorylates the intermediate filament protein Vimentin
Abstract
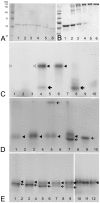
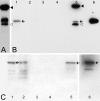
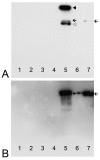
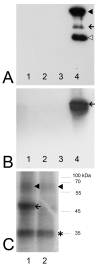
Background: Colocalization of Stk33 with vimentin by double immunofluorescence in certain cells indicated that vimentin might be a target for phosphorylation by the novel kinase Stk33. We therefore tested in vitro the ability of Stk33 to phosphorylate recombinant full length vimentin and amino-terminal truncated versions thereof. In order to prove that Stk33 and vimentin are also in vivo associated proteins co-immunoprecipitation experiments were carried out. For testing the enzymatic activity of immunoprecipitated Stk33 we incubated precipitated Stk33 with recombinant vimentin proteins. To investigate whether Stk33 binds directly to vimentin, an in vitro co-sedimentation assay was performed.
Results: The results of the kinase assays demonstrate that Stk33 is able to specifically phosphorylate the non-alpha-helical amino-terminal domain of vimentin in vitro. Furthermore, co-immunoprecipitation experiments employing cultured cell extracts indicate that Stk33 and vimentin are associated in vivo. Immunoprecipitated Stk33 has enzymatic activity as shown by successful phosphorylation of recombinant vimentin proteins. The results of the co-sedimentation assay suggest that vimentin binds directly to Stk33 and that no additional protein mediates the association.
Conclusion: We hypothesize that Stk33 is involved in the in vivo dynamics of the intermediate filament cytoskeleton by phosphorylating vimentin.
Figures

Similar articles
-
Co-localization of serine/threonine kinase 33 (Stk33) and vimentin in the hypothalamus.Cell Tissue Res. 2014 Jan;355(1):189-99. doi: 10.1007/s00441-013-1721-8. Epub 2013 Sep 22. Cell Tissue Res. 2014. PMID: 24057876
-
Domain-specific phosphorylation of vimentin and glial fibrillary acidic protein by PKN.Biochem Biophys Res Commun. 1997 May 29;234(3):621-5. doi: 10.1006/bbrc.1997.6669. Biochem Biophys Res Commun. 1997. PMID: 9175763
-
Aurora-B and Rho-kinase/ROCK, the two cleavage furrow kinases, independently regulate the progression of cytokinesis: possible existence of a novel cleavage furrow kinase phosphorylates ezrin/radixin/moesin (ERM).Genes Cells. 2005 Feb;10(2):127-37. doi: 10.1111/j.1365-2443.2005.00824.x. Genes Cells. 2005. PMID: 15676024
-
Differential expression pattern of the novel serine/threonine kinase, STK33, in mice and men.FEBS J. 2005 Oct;272(19):4884-98. doi: 10.1111/j.1742-4658.2005.04900.x. FEBS J. 2005. PMID: 16176263
-
Kinase activities of c-Mos and v-Mos proteins: a single amino acid exchange is responsible for constitutive activation of the 124 v-Mos kinase.Oncogene. 1995 Feb 16;10(4):623-30. Oncogene. 1995. PMID: 7862439
Cited by
-
Proteomic analysis of early HIV-1 nucleoprotein complexes.J Proteome Res. 2013 Feb 1;12(2):559-72. doi: 10.1021/pr300869h. Epub 2013 Jan 16. J Proteome Res. 2013. PMID: 23282062 Free PMC article.
-
HPD degradation regulated by the TTC36-STK33-PELI1 signaling axis induces tyrosinemia and neurological damage.Nat Commun. 2019 Sep 19;10(1):4266. doi: 10.1038/s41467-019-12011-0. Nat Commun. 2019. PMID: 31537781 Free PMC article.
-
Shared and Unique Disease Pathways in Amyotrophic Lateral Sclerosis and Parkinson's Disease Unveiled in Peripheral Blood Mononuclear Cells.ACS Chem Neurosci. 2023 Dec 6;14(23):4240-4251. doi: 10.1021/acschemneuro.3c00629. Epub 2023 Nov 8. ACS Chem Neurosci. 2023. PMID: 37939393 Free PMC article.
-
Role of serine/threonine kinase 33 methylation in colorectal cancer and its clinical significance.Oncol Lett. 2018 Feb;15(2):2153-2160. doi: 10.3892/ol.2017.7614. Epub 2017 Dec 14. Oncol Lett. 2018. PMID: 29434919 Free PMC article.
-
The coordinate cellular response to insulin-like growth factor-I (IGF-I) and insulin-like growth factor-binding protein-2 (IGFBP-2) is regulated through vimentin binding to receptor tyrosine phosphatase β (RPTPβ).J Biol Chem. 2015 May 1;290(18):11578-90. doi: 10.1074/jbc.M114.620237. Epub 2015 Mar 18. J Biol Chem. 2015. PMID: 25787077 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

