GATA4 is essential for jejunal function in mice
- PMID: 18812176
- PMCID: PMC2844802
- DOI: 10.1053/j.gastro.2008.07.074
GATA4 is essential for jejunal function in mice
Abstract
Background & aims: Although the zinc-finger transcription factor GATA4 has been implicated in regulating jejunal gene expression, the contribution of GATA4 in controlling jejunal physiology has not been addressed.
Methods: We generated mice in which the Gata4 gene was specifically deleted in the small intestinal epithelium. Measurements of plasma cholesterol and phospholipids, intestinal absorption of dietary fat and cholesterol, and gene expression were performed on these animals.
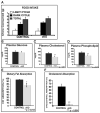
Results: Mice lacking GATA4 in the intestine displayed a dramatic block in their ability to absorb cholesterol and dietary fat. Comparison of the global gene expression profiles of control jejunum, control ileum, and GATA4 null jejunum by gene array analysis revealed that GATA4 null jejunum lost expression of 53% of the jejunal-specific gene set and gained expression of 47% of the set of genes unique to the ileum. These alterations in gene expression included a decrease in messenger RNAs (mRNAs) encoding lipid and cholesterol transporters as well as an increase in mRNAs encoding proteins involved in bile acid absorption.
Conclusions: Our data demonstrate that GATA4 is essential for jejunal function including fat and cholesterol absorption and confirm that GATA4 plays a pivotal role in determining jejunal vs ileal identity.
Conflict of interest statement
No conflicts of interest exist.
Figures




References
-
- Rubin DC. Intestinal morphogenesis. Curr Opin Gastroenterol. 2007;23:111–4. - PubMed
-
- van Wering HM, Bosse T, Musters A, et al. Complex regulation of the lactase-phlorizin hydrolase promoter by GATA-4. Am J Physiol Gastrointest Liver Physiol. 2004;287:G899–909. - PubMed
-
- Boudreau F, Rings EH, van Wering HM, et al. Hepatocyte nuclear factor-1 alpha, GATA-4, and caudal related homeodomain protein Cdx2 interact functionally to modulate intestinal gene transcription. Implication for the developmental regulation of the sucrase-isomaltase gene. J Biol Chem. 2002;277:31909–17. - PubMed
-
- Divine JK, Staloch LJ, Haveri H, et al. GATA-4, GATA-5, and GATA-6 activate the rat liver fatty acid binding protein gene in concert with HNF-1alpha. Am J Physiol Gastrointest Liver Physiol. 2004;287:G1086–99. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

