S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum
- PMID: 18812398
- PMCID: PMC2566862
- DOI: 10.1093/nar/gkn601
S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum
Abstract
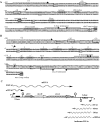
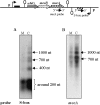
The ubiGmccBA operon of Clostridium acetobutylicum is involved in methionine to cysteine conversion. We showed that its expression is controlled by a complex regulatory system combining several RNA-based mechanisms. Two functional convergent promoters associated with transcriptional antitermination systems, a cysteine-specific T-box and an S-box riboswitch, are located upstream of and downstream from the ubiG operon, respectively. Several antisense RNAs were synthesized from the downstream S-box-dependent promoter, resulting in modulation of the level of ubiG transcript and of MccB activity. In contrast, the upstream T-box system did not appear to play a major role in regulation, leaving antisense transcription as the major regulatory mechanism for the ubiG operon. The abundance of sense and antisense transcripts was inversely correlated with the sulfur source availability. Deletion of the downstream promoter region completely abolished the sulfur-dependent control of the ubiG operon, and the expression of antisense transcripts in trans did not restore the regulation of the operon. Our data revealed important insights into the molecular mechanism of cis-antisense-mediated regulation, a control system only rarely observed in prokaryotes. We proposed a regulatory model in which the antisense RNA controlled the expression of the ubiG operon in cis via transcriptional interference at the ubiG locus.
Figures




References
-
- Guédon E, Martin-Verstraete I. In: Amino Acid Biosynthesis-pathways, Regulation and Metabolic Engineering. Wendisch VF, editor. Germany: Springer; 2007. pp. 195–218.
-
- Schelle MW, Bertozzi CR. Sulfate metabolism in mycobacteria. Chembiochem. 2006;7:1516–1524. - PubMed
-
- Sekowska A, Kung HF, Danchin A. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J. Mol. Microbiol. Biotechnol. 2000;2:145–177. - PubMed
-
- Greene RC. In: Escherichia coli and Salmonella, Cellular and Molecular Biology. Neidhardt FC, editor. Washington, DC: ASM Press; 1996. pp. 542–560.
-
- Ludwig ML, Matthews RG. Structure-based perspectives on B12-dependent enzymes. Annu. Rev. Biochem. 1997;66:269–313. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

