Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity
- PMID: 18812512
- PMCID: PMC2567449
- DOI: 10.1073/pnas.0803103105
Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity
Abstract
We outline a powerful method for the directed evolution of integral membrane proteins in the inner membrane of Escherichia coli. For a mammalian G protein-coupled receptor, we arrived at a sequence with an order-of-magnitude increase in functional expression that still retains the biochemical properties of wild type. This mutant also shows enhanced heterologous expression in eukaryotes (12-fold in Pichia pastoris and 3-fold in HEK293T cells) and greater stability when solubilized and purified, indicating that the biophysical properties of the protein had been under the pressure of selection. These improvements arise from multiple small contributions, which would be difficult to assemble by rational design. In a second screen, we rapidly pinpointed a single amino acid substitution in wild type that abolishes antagonist binding while retaining agonist-binding affinity. These approaches may alleviate existing bottlenecks in structural studies of these targets by providing sufficient quantities of stable variants in defined conformational states.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

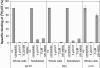


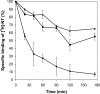
Comment in
-
Engineering G protein-coupled receptor expression in bacteria.Proc Natl Acad Sci U S A. 2008 Sep 30;105(39):14747-8. doi: 10.1073/pnas.0807741105. Epub 2008 Sep 23. Proc Natl Acad Sci U S A. 2008. PMID: 18812517 Free PMC article. No abstract available.
References
-
- Lundström K. Structural genomics of GPCRs. Trends Biotechnol. 2005;23:103–108. - PubMed
-
- Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. - PubMed
-
- Okada T, et al. X-ray diffraction analysis of three-dimensional crystals of bovine rhodopsin obtained from mixed micelles. J Struct Biol. 2000;130:73–80. - PubMed
-
- Rasmussen SG, et al. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

