Quantitation of Aurora kinase A gene copy number in urine sediments and bladder cancer detection
- PMID: 18812553
- PMCID: PMC2720731
- DOI: 10.1093/jnci/djn304
Quantitation of Aurora kinase A gene copy number in urine sediments and bladder cancer detection
Erratum in
- J Natl Cancer Inst. 2008 Dec 3;100(23):1740
Abstract
Background: Chromosome missegregation and the resulting aneuploidy is a common change in neoplasia. The Aurora kinase A (AURKA) gene, which encodes a key regulator of mitosis, is frequently amplified and/or overexpressed in cancer cells, and the level of AURKA amplification is associated with the level of aneuploidy. We examined whether AURKA gene amplification is a biomarker for the detection of bladder cancer.
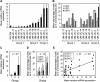
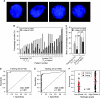
Methods: The effect of ectopic expression of Aurora kinase A (AURKA) using an adenoviral vector in simian virus 40-immortalized urothelial cells (SV-HUC) on centrosome multiplication and chromosome copy number was measured in vitro by immunofluorescence and fluorescence in situ hybridization (FISH), respectively. The FISH test was also used to examine AURKA gene copy number in exfoliated cells in voided urine samples from 23 patients with bladder cancer and 7 healthy control subjects (training set), generating a model for bladder cancer detection that was subsequently validated in an independent set of voided urine samples from 100 bladder cancer patients and 148 control subjects (92 healthy individuals and 56 patients with benign urologic disorders). An AURKA gene score (the proportion of cells with three or more AURKA signals) was used to produce receiver operating characteristic (ROC) curves and to calculate the specificity and sensitivity of the AURKA FISH test. Differences between mean AURKA scores in different pathogenetic groups of bladder cancer stratified according to histological grade and stage were tested by unpaired Mann-Whitney t tests or one-way Wilcoxon tests. All statistical tests were two-sided.
Results: Forced overexpression of AURKA in urothelial cells induced amplification of centrosomes, chromosome missegregation, and aneuploidy, and natural overexpression was detectable in in situ lesions from patients with bladder cancer. The FISH test for the AURKA gene copy number performed on the validation set yielded a specificity of 96.6% (95% confidence interval [CI] = 92.3% to 98.5%) and sensitivity of 87% (95% CI = 79.0% to 92.2%) and an area under the ROC curve of 0.939 (95% CI = 0.906 to 0.971; P < .001).
Conclusion: Overexpression of AURKA can cause aneuploidy in urothelial cells, and the AURKA gene copy number is a promising biomarker for detection of bladder cancer.
Figures


References
-
- Perkins AS, Stern DF. Molecular Biology of Cancer. 5th ed. Philadelphia, PA: Lippincott-Raven Publishers; 1997.
-
- Heim S, Mitelman F. Cancer Cytogenetics. 2nd ed. New York: Wiley-Liss; 1995.
-
- Sen S. Aneuploidy and cancer. Curr Opin Oncol. 2000;12(1):82–88. - PubMed
-
- Zhou H, Kuang J, Zhong L, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20(2):189–193. - PubMed
-
- Katayama H, Zhou H, Li Q, Tatsuka M, Sen S. Interaction and feedback regulation between STK15/BTAK/Aurora-A kinase and protein phosphatase 1 through mitotic cell division cycle. J Biol Chem. 2001;276(49):46219–46224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

