Analysis of the Pseudoalteromonas tunicata genome reveals properties of a surface-associated life style in the marine environment
- PMID: 18813346
- PMCID: PMC2536512
- DOI: 10.1371/journal.pone.0003252
Analysis of the Pseudoalteromonas tunicata genome reveals properties of a surface-associated life style in the marine environment
Abstract
Background: Colonisation of sessile eukaryotic host surfaces (e.g. invertebrates and seaweeds) by bacteria is common in the marine environment and is expected to create significant inter-species competition and other interactions. The bacterium Pseudoalteromonas tunicata is a successful competitor on marine surfaces owing primarily to its ability to produce a number of inhibitory molecules. As such P. tunicata has become a model organism for the studies into processes of surface colonisation and eukaryotic host-bacteria interactions.
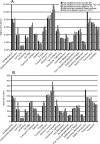
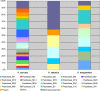
Methodology/principal findings: To gain a broader understanding into the adaptation to a surface-associated life-style, we have sequenced and analysed the genome of P. tunicata and compared it to the genomes of closely related strains. We found that the P. tunicata genome contains several genes and gene clusters that are involved in the production of inhibitory compounds against surface competitors and secondary colonisers. Features of P. tunicata's oxidative stress response, iron scavenging and nutrient acquisition show that the organism is well adapted to high-density communities on surfaces. Variation of the P. tunicata genome is suggested by several landmarks of genetic rearrangements and mobile genetic elements (e.g. transposons, CRISPRs, phage). Surface attachment is likely to be mediated by curli, novel pili, a number of extracellular polymers and potentially other unexpected cell surface proteins. The P. tunicata genome also shows a utilisation pattern of extracellular polymers that would avoid a degradation of its recognised hosts, while potentially causing detrimental effects on other host types. In addition, the prevalence of recognised virulence genes suggests that P. tunicata has the potential for pathogenic interactions.
Conclusions/significance: The genome analysis has revealed several physiological features that would provide P. tunciata with competitive advantage against other members of the surface-associated community. We have also identified properties that could mediate interactions with surfaces other than its currently recognised hosts. This together with the detection of known virulence genes leads to the hypothesis that P. tunicata maintains a carefully regulated balance between beneficial and detrimental interactions with a range of host surfaces.
Conflict of interest statement
Figures



References
-
- Braithwaite RA, McEvoy LA. Marine biofouling on fish farms and its remediation. Adv Mar Biol. 2005;47:215–252. - PubMed
-
- Berk SG, Mitchell R, Bobbie RJ, Nickels JS, White DC. Microfouling on metal surfaces exposed to seawater. International Biodeteriation and Biodegradation. 2001;34:387–399.
-
- Qian PY, Lau S, Dahms HU, Dobretsov S, Harder T. Marine Biofilms as Mediators of Colonization by Marine Macroorganisms: Implications for Antifouling and Aquaculture. Mar Biotechnol. 2007;9:399–410. - PubMed
-
- Egan S, Thomas T, Kjelleberg S. Unlocking the diversity and biotechnological potential of marine surface associated microbial communities. Curr Opinion Microbiol. 2008;11:219–225. - PubMed
-
- Wang Y, Tang XX, Yang Z, Yu ZM. Effect of alginic acid decomposing bacterium on the growth of Laminaria japonica (Phaeophyceae). J Environ Sci. 2006;18:543–551. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

