Epstein-Barr virus latent membrane protein 2A exploits Notch1 to alter B-cell identity in vivo
- PMID: 18815281
- PMCID: PMC2614628
- DOI: 10.1182/blood-2008-06-160937
Epstein-Barr virus latent membrane protein 2A exploits Notch1 to alter B-cell identity in vivo
Abstract
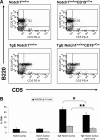
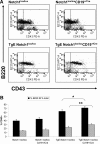
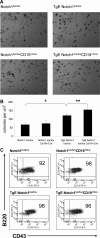
Expression of latent membrane protein 2 (LMP2A) during B-cell development leads to global alterations in gene transcription similar to those seen in Hodgkin Reed-Sternberg cells of Hodgkin lymphoma (HL). Along with the consistent detection of LMP2A in Epstein-Barr virus-associated HL, this implicates a role for LMP2A in the pathogenesis of HL. We have shown that LMP2A constitutively activates the Notch1 pathway to autoregulate the LMP2A promoter. To determine whether constitutive activation of the Notch pathway is important for LMP2A-mediated alterations in B-cell development in vivo, TgE-LMP2A-transgenic mice were intercrossed with mice expressing loxP-flanked Notch1 genes and Cre recombinase. B cells from TgE Notch1(lox/lox)-CD19(+/Cre) mice have an increase in immunoglobulin M and CD43 and a decrease in CD5 expression in the bone marrow compared with TgE Notch1(lox/lox) mice, indicating the LMP2A signal for developmental aberrations is impaired in the absence of Notch1. Real-time reverse-transcribed polymerase chain reaction analysis reveals that LMP2A requires the Notch1 pathway to alter levels of B cell-specific transcription factors, E2A and EBF. Interestingly, Notch1 appears to be important for LMP2A-mediated survival in low interleukin-7. We propose that LMP2A and the Notch1 pathway may cooperate to induce the alterations in B-cell identity seen in Hodgkin Reed-Sternberg cells.
Figures




References
-
- Rickinson AB, Kieff E. Epstein-Barr Virus. Philadelphia, PA: Lippincott-Raven Publishers; 2007. pp. 2397–2446.
-
- Caldwell RG, Wilson JB, Anderson SJ, Longnecker R. Epstein-Barr virus LMP2A drives B-cell development and survival in the absence of normal B cell receptor signals. Immunity. 1998;9:405–411. - PubMed
-
- Casola S, Otipoby KL, Alimzhanov M, et al. B cell receptor signal strength determines B cell fate. Nat Immunol. 2004;5:317–327. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

