Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant
- PMID: 18815367
- PMCID: PMC2567475
- DOI: 10.1073/pnas.0808066105
Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant
Abstract
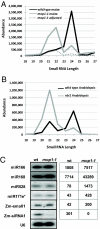
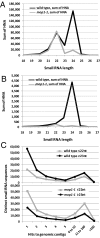
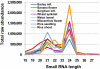
Small RNAs from plants are known to be highly complex and abundant, with this complexity proportional to genome size. Most endogenous siRNAs in Arabidopsis are dependent on RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) for their biogenesis. Recent work has demonstrated that the maize MEDIATOR OF PARAMUTATION1 (mop1) gene is a predicted ortholog of RDR2. The mop1 gene is required for establishment of paramutation and maintenance of transcriptional silencing of transposons and transgenes, suggesting the potential involvement of small RNAs. We analyzed small RNAs in wild-type maize and in the isogenic mop1-1 loss-of-function mutant by using Illumina's sequencing-by-synthesis (SBS) technology, which allowed us to characterize the complement of maize small RNAs to considerable depth. Similar to rdr2 in Arabidopsis, in mop1-1, the 24-nucleotide (nt) endogenous heterochromatic short-interfering siRNAs were dramatically reduced, resulting in an enrichment of miRNAs and transacting siRNAs. In contrast to the Arabidopsis rdr2 mutant, the mop1-1 plants retained a highly abundant heterochromatic approximately 22-nt class of small RNAs, suggesting a second mechanism for heterochromatic siRNA production. The enrichment of miRNAs and loss of 24-nt heterochromatic siRNAs in mop1-1 should be advantageous for miRNA discovery as the maize genome becomes more fully sequenced.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15:1494–1500. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

