Presenting and exploring biological pathways with PathVisio
- PMID: 18817533
- PMCID: PMC2569944
- DOI: 10.1186/1471-2105-9-399
Presenting and exploring biological pathways with PathVisio
Abstract
Background: Biological pathways are a useful abstraction of biological concepts, and software tools to deal with pathway diagrams can help biological research. PathVisio is a new visualization tool for biological pathways that mimics the popular GenMAPP tool with a completely new Java implementation that allows better integration with other open source projects. The GenMAPP MAPP file format is replaced by GPML, a new XML file format that provides seamless exchange of graphical pathway information among multiple programs.
Results: PathVisio can be combined with other bioinformatics tools to open up three possible uses: visual compilation of biological knowledge, interpretation of high-throughput expression datasets, and computational augmentation of pathways with interaction information. PathVisio is open source software and available at http://www.pathvisio.org.
Conclusion: PathVisio is a graphical editor for biological pathways, with flexibility and ease of use as primary goals.
Figures
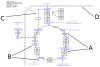

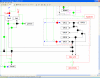
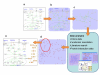
References
-
- BioPAX wiki http://www.biopax.org/
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

