Structures of human cytochrome P-450 2E1. Insights into the binding of inhibitors and both small molecular weight and fatty acid substrates
- PMID: 18818195
- PMCID: PMC2586265
- DOI: 10.1074/jbc.M805999200
Structures of human cytochrome P-450 2E1. Insights into the binding of inhibitors and both small molecular weight and fatty acid substrates
Abstract
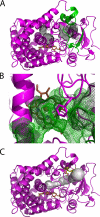
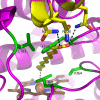
Human microsomal cytochrome P-450 2E1 (CYP2E1) monooxygenates > 70 low molecular weight xenobiotic compounds, as well as much larger endogenous fatty acid signaling molecules such as arachidonic acid. In the process, CYP2E1 can generate toxic or carcinogenic compounds, as occurs with acetaminophen overdose, nitrosamines in cigarette smoke, and reactive oxygen species from uncoupled catalysis. Thus, the diverse roles that CYP2E1 has in normal physiology, toxicity, and drug metabolism are related to its ability to metabolize diverse classes of ligands, but the structural basis for this was previously unknown. Structures of human CYP2E1 have been solved to 2.2 angstroms for an indazole complex and 2.6 angstroms for a 4-methylpyrazole complex. Both inhibitors bind to the heme iron and hydrogen bond to Thr303 within the active site. Complementing its small molecular weight substrates, the hydrophobic CYP2E1 active site is the smallest yet observed for a human cytochrome P-450. The CYP2E1 active site also has two adjacent voids: one enclosed above the I helix and the other forming a channel to the protein surface. Minor repositioning of the Phe478 aromatic ring that separates the active site and access channel would allow the carboxylate of fatty acid substrates to interact with conserved 216QXXNN220 residues in the access channel while positioning the hydrocarbon terminus in the active site, consistent with experimentally observed omega-1 hydroxylation of saturated fatty acids. Thus, these structures provide insights into the ability of CYP2E1 to effectively bind and metabolize both small molecule substrates and fatty acids.
Figures




References
-
- Ioannides, C., and Lewis, D. F. (2004) Curr. Top. Med. Chem. 4 1767-1788 - PubMed
-
- Bieche, I., Narjoz, C., Asselah, T., Vacher, S., Marcellin, P., Lidereau, R., Beaune, P., and de Waziers, I. (2007) Pharmacogenet. Genomics 17 731-742 - PubMed
-
- Shimada, T., Yamazaki, H., Mimura, M., Inui, Y., and Guengerich, F. P. (1994) J. Pharmacol. Exp. Ther. 270 414-423 - PubMed
-
- Ding, X., and Kaminsky, L. S. (2003) Annu. Rev. Pharmacol. Toxicol. 43 149-173 - PubMed
-
- Nanji, A. A., Zhao, S., Sadrzadeh, S. M., Dannenberg, A. J., Tahan, S. R., and Waxman, D. J. (1994) Alcohol. Clin. Exp. Res. 18 1280-1285 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

