On the mechanism of pore formation by melittin
- PMID: 18819911
- PMCID: PMC2662212
- DOI: 10.1074/jbc.M805171200
On the mechanism of pore formation by melittin
Abstract
The mechanism of pore formation of lytic peptides, such as melittin from bee venom, is thought to involve binding to the membrane surface, followed by insertion at threshold levels of bound peptide. We show that in membranes composed of zwitterionic lipids, i.e. phosphatidylcholine, melittin not only forms pores but also inhibits pore formation. We propose that these two modes of action are the result of two competing reactions: direct insertion into the membrane and binding parallel to the membrane surface. The direct insertion of melittin leads to pore formation, whereas the parallel conformation is inactive and prevents other melittin molecules from inserting, hence preventing pore formation.
Figures

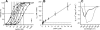

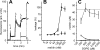
Similar articles
-
The synergistic action of melittin and phospholipase A2 with lipid membranes: development of linear dichroism for membrane-insertion kinetics.Protein Pept Lett. 2010 Nov;17(11):1351-62. doi: 10.2174/0929866511009011351. Protein Pept Lett. 2010. PMID: 20673225
-
Modulation of tryptophan environment in membrane-bound melittin by negatively charged phospholipids: implications in membrane organization and function.Biochemistry. 1997 Nov 25;36(47):14291-305. doi: 10.1021/bi971933j. Biochemistry. 1997. PMID: 9398147
-
Barrel-stave model or toroidal model? A case study on melittin pores.Biophys J. 2001 Sep;81(3):1475-85. doi: 10.1016/S0006-3495(01)75802-X. Biophys J. 2001. PMID: 11509361 Free PMC article.
-
How Melittin Inserts into Cell Membrane: Conformational Changes, Inter-Peptide Cooperation, and Disturbance on the Membrane.Molecules. 2019 May 7;24(9):1775. doi: 10.3390/molecules24091775. Molecules. 2019. PMID: 31067828 Free PMC article. Review.
-
Model Membrane and Cell Studies of Antimicrobial Activity of Melittin Analogues.Curr Top Med Chem. 2016;16(1):40-5. doi: 10.2174/1568026615666150703115919. Curr Top Med Chem. 2016. PMID: 26139117 Review.
Cited by
-
Hybrid bilayer membranes on metallurgical polished aluminum.Sci Rep. 2021 May 6;11(1):9648. doi: 10.1038/s41598-021-89150-2. Sci Rep. 2021. PMID: 33958658 Free PMC article.
-
Gain-of-function analogues of the pore-forming peptide melittin selected by orthogonal high-throughput screening.J Am Chem Soc. 2012 Aug 1;134(30):12732-41. doi: 10.1021/ja3042004. Epub 2012 Jul 18. J Am Chem Soc. 2012. PMID: 22731650 Free PMC article.
-
Enhanced Therapeutic Effect of Optimized Melittin-dKLA, a Peptide Agent Targeting M2-like Tumor-Associated Macrophages in Triple-Negative Breast Cancer.Int J Mol Sci. 2022 Dec 12;23(24):15751. doi: 10.3390/ijms232415751. Int J Mol Sci. 2022. PMID: 36555393 Free PMC article.
-
Tuneable poration: host defense peptides as sequence probes for antimicrobial mechanisms.Sci Rep. 2018 Oct 8;8(1):14926. doi: 10.1038/s41598-018-33289-y. Sci Rep. 2018. PMID: 30297841 Free PMC article.
-
Reassessing the Host Defense Peptide Landscape.Front Chem. 2019 Feb 4;7:43. doi: 10.3389/fchem.2019.00043. eCollection 2019. Front Chem. 2019. PMID: 30778385 Free PMC article. Review.
References
-
- Brogden, K. A. (2005) Nat. Rev. Microbiol. 3 238–250 - PubMed
-
- Finlay, B. B., and Hancock, R. E. (2004) Nat. Rev. Microbiol. 2 497–504 - PubMed
-
- Raghuraman, H., and Chattopadhyay, A. (2007) Biosci. Rep. 27 189–223 - PubMed
-
- Son, D. J., Lee, J. W., Lee, Y. H., Song, H. S., Lee, C. K., and Hong, J. T. (2007) Pharmacol. Ther. 115 246–270 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

