Nuclear export competence of pre-40S subunits in fission yeast requires the ribosomal protein Rps2
- PMID: 18820293
- PMCID: PMC2577348
- DOI: 10.1093/nar/gkn625
Nuclear export competence of pre-40S subunits in fission yeast requires the ribosomal protein Rps2
Abstract
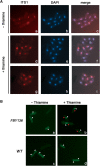
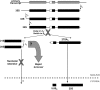
Ribosome biogenesis is an evolutionarily conserved pathway that requires ribosomal and nonribosomal proteins. Here, we investigated the role of the ribosomal protein S2 (Rps2) in fission yeast ribosome synthesis. As for many budding yeast ribosomal proteins, Rps2 was essential for cell viability in fission yeast and the genetic depletion of Rps2 caused a complete inhibition of 40S ribosomal subunit production. The pattern of pre-rRNA processing upon depletion of Rps2 revealed a reduction of 27SA(2) pre-rRNAs and the concomitant production of 21S rRNA precursors, consistent with a role for Rps2 in efficient cleavage at site A(2) within the 32S pre-rRNA. Importantly, kinetics of pre-rRNA accumulation as determined by rRNA pulse-chases assays indicated that a small fraction of 35S precursors matured into 20S-containing particles, suggesting that most 40S precursors were rapidly degraded in the absence of Rps2. Analysis of steady-state RNA levels revealed that some pre-40S particles were produced in Rps2-depleted cells, but that these precursors were retained in the nucleolus. Our findings suggest a role for Rps2 in a mechanism that monitors pre-40S export competence.
Figures
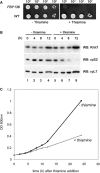
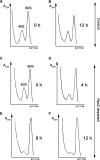

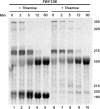
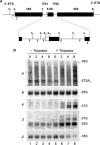
Similar articles
-
Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae.Mol Microbiol. 2004 Apr;52(1):141-58. doi: 10.1111/j.1365-2958.2003.03973.x. Mol Microbiol. 2004. PMID: 15049817
-
Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein.EMBO J. 2001 Aug 1;20(15):4204-13. doi: 10.1093/emboj/20.15.4204. EMBO J. 2001. PMID: 11483523 Free PMC article.
-
Sdo1p, the yeast orthologue of Shwachman-Bodian-Diamond syndrome protein, binds RNA and interacts with nuclear rRNA-processing factors.Yeast. 2009 May;26(5):287-98. doi: 10.1002/yea.1668. Yeast. 2009. PMID: 19350533
-
Nuclear export and cytoplasmic maturation of ribosomal subunits.FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
-
Ribosomal RNA processing and ribosome biogenesis in eukaryotes.IUBMB Life. 2004 Aug;56(8):457-65. doi: 10.1080/15216540400010867. IUBMB Life. 2004. PMID: 15545225 Review.
Cited by
-
Rmt1 catalyzes zinc-finger independent arginine methylation of ribosomal protein Rps2 in Saccharomyces cerevisiae.Biochem Biophys Res Commun. 2010 Jan 22;391(4):1658-62. doi: 10.1016/j.bbrc.2009.12.112. Epub 2009 Dec 24. Biochem Biophys Res Commun. 2010. PMID: 20035717 Free PMC article.
-
Discovery of peptidylarginine deiminase-4 substrates by protein array: antagonistic citrullination and methylation of human ribosomal protein S2.Mol Biosyst. 2011 Jul;7(7):2286-95. doi: 10.1039/c1mb05089c. Epub 2011 May 16. Mol Biosyst. 2011. PMID: 21584310 Free PMC article.
-
The 40S ribosomal protein uS5 (RPS2) assembles into an extraribosomal complex with human ZNF277 that competes with the PRMT3-uS5 interaction.J Biol Chem. 2019 Feb 8;294(6):1944-1955. doi: 10.1074/jbc.RA118.004928. Epub 2018 Dec 10. J Biol Chem. 2019. PMID: 30530495 Free PMC article.
-
A methyltransferase-independent function for Rmt3 in ribosomal subunit homeostasis.J Biol Chem. 2009 May 29;284(22):15026-37. doi: 10.1074/jbc.M109.004812. Epub 2009 Apr 9. J Biol Chem. 2009. PMID: 19359250 Free PMC article.
-
The conserved RNA-binding protein Seb1 promotes cotranscriptional ribosomal RNA processing by controlling RNA polymerase I progression.Nat Commun. 2023 May 25;14(1):3013. doi: 10.1038/s41467-023-38826-6. Nat Commun. 2023. PMID: 37230993 Free PMC article.
References
-
- Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. - PubMed
-
- Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

