The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor
- PMID: 18820679
- PMCID: PMC2596276
- DOI: 10.1038/nature07405
The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor
Abstract
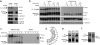
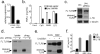
Mammalian Toll-like receptors (TLRs) 3, 7, 8 and 9 initiate immune responses to infection by recognizing microbial nucleic acids; however, these responses come at the cost of potential autoimmunity owing to inappropriate recognition of self nucleic acids. The localization of TLR9 and TLR7 to intracellular compartments seems to have a role in facilitating responses to viral nucleic acids while maintaining tolerance to self nucleic acids, yet the cell biology regulating the transport and localization of these receptors remains poorly understood. Here we define the route by which TLR9 and TLR7 exit the endoplasmic reticulum and travel to endolysosomes in mouse macrophages and dendritic cells. The ectodomains of TLR9 and TLR7 are cleaved in the endolysosome, such that no full-length protein is detectable in the compartment where ligand is recognized. Notably, although both the full-length and cleaved forms of TLR9 are capable of binding ligand, only the processed form recruits MyD88 on activation, indicating that this truncated receptor, rather than the full-length form, is functional. Furthermore, conditions that prevent receptor proteolysis, including forced TLR9 surface localization, render the receptor non-functional. We propose that ectodomain cleavage represents a strategy to restrict receptor activation to endolysosomal compartments and prevent TLRs from responding to self nucleic acids.
Figures


References
-
- Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. - PubMed
-
- Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. - PubMed
-
- Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. - PubMed
-
- Latz E, et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

