A genome-wide association study for late-onset Alzheimer's disease using DNA pooling
- PMID: 18823527
- PMCID: PMC2570675
- DOI: 10.1186/1755-8794-1-44
A genome-wide association study for late-onset Alzheimer's disease using DNA pooling
Abstract
Background: Late-onset Alzheimer's disease (LOAD) is an age related neurodegenerative disease with a high prevalence that places major demands on healthcare resources in societies with increasingly aged populations. The only extensively replicable genetic risk factor for LOAD is the apolipoprotein E gene. In order to identify additional genetic risk loci we have conducted a genome-wide association (GWA) study in a large LOAD case - control sample, reducing costs through the use of DNA pooling.
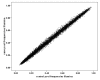
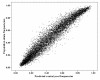
Methods: DNA samples were collected from 1,082 individuals with LOAD and 1,239 control subjects. Age at onset ranged from 60 to 95 and Controls were matched for age (mean = 76.53 years, SD = 33), gender and ethnicity. Equimolar amounts of each DNA sample were added to either a case or control pool. The pools were genotyped using Illumina HumanHap300 and Illumina Sentrix HumanHap240S arrays testing 561,494 SNPs. 114 of our best hit SNPs from the pooling data were identified and then individually genotyped in the case - control sample used to construct the pools.
Results: Highly significant association with LOAD was observed at the APOE locus confirming the validity of the pooled genotyping approach.For 109 SNPs outside the APOE locus, we obtained uncorrected p-values </= 0.05 for 74 after individual genotyping. To further test these associations, we added control data from 1400 subjects from the 1958 Birth Cohort with the evidence for association increasing to 3.4 x 10-6 for our strongest finding, rs727153.rs727153 lies 13 kb from the start of transcription of lecithin retinol acyltransferase (phosphatidylcholine - retinol O-acyltransferase, LRAT). Five of seven tag SNPs chosen to cover LRAT showed significant association with LOAD with a SNP in intron 2 of LRAT, showing greatest evidence of association (rs201825, p-value = 6.1 x 10-7).
Conclusion: We have validated the pooling method for GWA studies by both identifying the APOE locus and by observing a strong enrichment for significantly associated SNPs. We provide evidence for LRAT as a novel candidate gene for LOAD. LRAT plays a prominent role in the Vitamin A cascade, a system that has been previously implicated in LOAD.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

