iRefIndex: a consolidated protein interaction database with provenance
- PMID: 18823568
- PMCID: PMC2573892
- DOI: 10.1186/1471-2105-9-405
iRefIndex: a consolidated protein interaction database with provenance
Abstract
Background: Interaction data for a given protein may be spread across multiple databases. We set out to create a unifying index that would facilitate searching for these data and that would group together redundant interaction data while recording the methods used to perform this grouping.
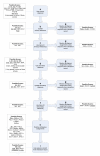
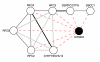
Results: We present a method to generate a key for a protein interaction record and a key for each participant protein. These keys may be generated by anyone using only the primary sequence of the proteins, their taxonomy identifiers and the Secure Hash Algorithm. Two interaction records will have identical keys if they refer to the same set of identical protein sequences and taxonomy identifiers. We define records with identical keys as a redundant group. Our method required that we map protein database references found in interaction records to current protein sequence records. Operations performed during this mapping are described by a mapping score that may provide valuable feedback to source interaction databases on problematic references that are malformed, deprecated, ambiguous or unfound. Keys for protein participants allow for retrieval of interaction information independent of the protein references used in the original records.
Conclusion: We have applied our method to protein interaction records from BIND, BioGrid, DIP, HPRD, IntAct, MINT, MPact, MPPI and OPHID. The resulting interaction reference index is provided in PSI-MITAB 2.5 format at http://irefindex.uio.no. This index may form the basis of alternative redundant groupings based on gene identifiers or near sequence identity groupings.
Figures


References
-
- Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, Tyers M, Salama JJ, Moore S, Ceol A, Chatr-Aryamontri A, Oesterheld M, Stumpflen V, Salwinski L, Nerothin J, Cerami E, Cusick ME, Vidal M, Gilson M, Armstrong J, Woollard P, Hogue C, Eisenberg D, Cesareni G, Apweiler R, Hermjakob H. Broadening the horizon–level 2.5 of the HUPO-PSI format for molecular interactions. BMC biology. 2007;5:44. doi: 10.1186/1741-7007-5-44. - DOI - PMC - PubMed
-
- INSDC: International Nucleotide Sequence Database Collaboration http://www.insdc.org
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

