The spindle assembly checkpoint is satisfied in the absence of interkinetochore tension during mitosis with unreplicated genomes
- PMID: 18824563
- PMCID: PMC2557037
- DOI: 10.1083/jcb.200801038
The spindle assembly checkpoint is satisfied in the absence of interkinetochore tension during mitosis with unreplicated genomes
Abstract
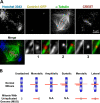
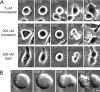
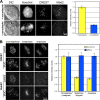
The accuracy of chromosome segregation is enhanced by the spindle assembly checkpoint (SAC). The SAC is thought to monitor two distinct events: attachment of kinetochores to microtubules and the stretch of the centromere between the sister kinetochores that arises only when the chromosome becomes properly bioriented. We examined human cells undergoing mitosis with unreplicated genomes (MUG). Kinetochores in these cells are not paired, which implies that the centromere cannot be stretched; however, cells progress through mitosis. A SAC is present during MUG as cells arrest in response to nocodazole, taxol, or monastrol treatments. Mad2 is recruited to unattached MUG kinetochores and released upon their attachment. In contrast, BubR1 remains on attached kinetochores and exhibits a level of phosphorylation consistent with the inability of MUG spindles to establish normal levels of centromere tension. Thus, kinetochore attachment to microtubules is sufficient to satisfy the SAC even in the absence of interkinetochore tension.
Figures


References
-
- Balczon, R.C. 2001. Overexpression of cyclin A in human HeLa cells induces detachment of kinetochores and spindle pole/centrosome overproduction. Chromosoma. 110:381–392. - PubMed
-
- Brinkley, B.R., R.P. Zinkowski, W.L. Mollon, F.M. Davis, M.A. Pisegna, M. Pershouse, and P.N. Rao. 1988. Movement and segregation of kinetochores experimentally detached from mammalian chromosomes. Nature. 336:251–254. - PubMed
-
- Ganem, N.J., K. Upton, and D.A. Compton. 2005. Efficient mitosis in human cells lacking polewards microtubule flux. Curr. Biol. 15:1827–1832. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

