A 106-kDa aminopeptidase is a putative receptor for Bacillus thuringiensis Cry11Ba toxin in the mosquito Anopheles gambiae
- PMID: 18826260
- PMCID: PMC2660596
- DOI: 10.1021/bi801181g
A 106-kDa aminopeptidase is a putative receptor for Bacillus thuringiensis Cry11Ba toxin in the mosquito Anopheles gambiae
Abstract
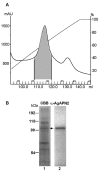
Bacillus thuringiensis (Bt) insecticidal toxins bind to receptors on midgut epithelial cells of susceptible insects, and binding triggers biochemical events that lead to insect mortality. Recently, a 100-kDa aminopeptidase N (APN) was isolated from brush border membrane vesicles (BBMV) of Anopheles quadrimaculatus and shown to bind Cry11Ba toxin with surface plasmon resonance (SPR) detection [Abdullah et al. (2006) BMC Biochem. 7, 16]. In our study, a 106-kDa APN, called AgAPN2, released by phosphatidylinositol-specific phospholipase C (PI-PLC) from Anopheles gambiae BBMV was extracted by Cry11Ba bound to beads. The AgAPN2 cDNA was cloned, and analysis of the predicted AgAPN2 protein revealed a zinc-binding motif (HEIAH), three potential N-glycosylation sites, and a predicted glycosylphosphatidylinositol (GPI) anchor site. Immunohistochemistry localized AgAPN2 to the microvilli of the posterior midgut. A 70-kDa fragment of the 106-kDa APN was expressed in Escherichia coli. When purified, it competitively displaced 125I-Cry11Ba binding to An. gambiae BBMV and bound Cry11Ba on dot blot and microtiter plate binding assays with a calculated K d of 6.4 nM. Notably, this truncated peptide inhibited Cry11Ba toxicity to An. gambiae larvae. These results are evidence that the 106-kDa GPI-anchored APN is a specific binding protein, and a putative midgut receptor, for Bt Cry11Ba toxin.
Figures



Similar articles
-
Anopheles gambiae alkaline phosphatase is a functional receptor of Bacillus thuringiensis jegathesan Cry11Ba toxin.Biochemistry. 2009 Oct 20;48(41):9785-93. doi: 10.1021/bi9014538. Biochemistry. 2009. PMID: 19747003
-
AgCad2 cadherin in Anopheles gambiae larvae is a putative receptor of Cry11Ba toxin of Bacillus thuringiensis subsp. jegathesan.Insect Biochem Mol Biol. 2013 Feb;43(2):153-61. doi: 10.1016/j.ibmb.2012.11.007. Epub 2012 Dec 8. Insect Biochem Mol Biol. 2013. PMID: 23231770
-
Analyses of α-amylase and α-glucosidase in the malaria vector mosquito, Anopheles gambiae, as receptors of Cry11Ba toxin of Bacillus thuringiensis subsp. jegathesan.Insect Biochem Mol Biol. 2013 Oct;43(10):907-15. doi: 10.1016/j.ibmb.2013.07.003. Epub 2013 Jul 18. Insect Biochem Mol Biol. 2013. PMID: 23872242
-
[Advances in receptor-mediated resistance mechanisms of Lepidopteran insects to Bacillus thuringiensis toxin].Sheng Wu Gong Cheng Xue Bao. 2022 May 25;38(5):1809-1823. doi: 10.13345/j.cjb.210834. Sheng Wu Gong Cheng Xue Bao. 2022. PMID: 35611730 Review. Chinese.
-
Bacillus thuringiensis subsp. israelensis and its dipteran-specific toxins.Toxins (Basel). 2014 Mar 28;6(4):1222-43. doi: 10.3390/toxins6041222. Toxins (Basel). 2014. PMID: 24686769 Free PMC article. Review.
Cited by
-
Two specific membrane-bound aminopeptidase N isoforms from Aedes aegypti larvae serve as functional receptors for the Bacillus thuringiensis Cry4Ba toxin implicating counterpart specificity.Biochem Biophys Res Commun. 2015 May 29;461(2):300-6. doi: 10.1016/j.bbrc.2015.04.026. Epub 2015 Apr 12. Biochem Biophys Res Commun. 2015. PMID: 25871797 Free PMC article.
-
Loop residues of the receptor binding domain of Bacillus thuringiensis Cry11Ba toxin are important for mosquitocidal activity.FEBS Lett. 2009 Jun 18;583(12):2021-30. doi: 10.1016/j.febslet.2009.05.020. Epub 2009 May 18. FEBS Lett. 2009. PMID: 19450583 Free PMC article.
-
Bacillus thuringiensis toxins: an overview of their biocidal activity.Toxins (Basel). 2014 Dec 11;6(12):3296-325. doi: 10.3390/toxins6123296. Toxins (Basel). 2014. PMID: 25514092 Free PMC article. Review.
-
A 104 kDa Aedes aegypti aminopeptidase N is a putative receptor for the Cry11Aa toxin from Bacillus thuringiensis subsp. israelensis.Insect Biochem Mol Biol. 2013 Dec;43(12):1201-8. doi: 10.1016/j.ibmb.2013.09.007. Epub 2013 Oct 12. Insect Biochem Mol Biol. 2013. PMID: 24128608 Free PMC article.
-
Cadherin fragments from Anopheles gambiae synergize Bacillus thuringiensis Cry4Ba's toxicity against Aedes aegypti larvae.Appl Environ Microbiol. 2009 Nov;75(22):7280-2. doi: 10.1128/AEM.01870-09. Epub 2009 Oct 2. Appl Environ Microbiol. 2009. PMID: 19801487 Free PMC article.
References
-
- Walker K, Lynch M. Contributions of Anopheles larval control to malaria suppression in tropical Africa: review of achievements and potential. Med Vet Entomol. 2007;21:2–21. - PubMed
-
- Federici BA, Park HW, Bideshi DK, Wirth MC, Johnson JJ, Sakano Y, Tang M. Developing recombinant bacteria for control of mosquito larvae. J Am Mosq Control Assoc. 2007;23:164–175. - PubMed
-
- Bravo A, Gomez I, Conde J, Munoz-Garay C, Sanchez J, Miranda R, Zhuang M, Gill SS, Soberon M. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochim Biophys Acta. 2004;1667:38–46. - PubMed
-
- Ravoahangimalala O, Charles JF. In vitro binding of Bacillus thuringiensis var. israelensis individual toxins to midgut cells of Anopheles gambiae larvae (Diptera: Culicidae) FEBS Lett. 1995;362:111–115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

