Single-particle kinetics of influenza virus membrane fusion
- PMID: 18829437
- PMCID: PMC2556630
- DOI: 10.1073/pnas.0807771105
Single-particle kinetics of influenza virus membrane fusion
Abstract
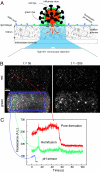
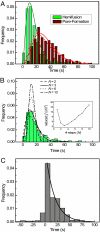
Membrane fusion is an essential step during entry of enveloped viruses into cells. Conventional fusion assays are generally limited to observation of ensembles of multiple fusion events, confounding more detailed analysis of the sequence of the molecular steps involved. We have developed an in vitro, two-color fluorescence assay to monitor kinetics of single virus particles fusing with a target bilayer on an essentially fluid support. Analysis of lipid- and content-mixing trajectories on a particle-by-particle basis provides evidence for multiple, long-lived kinetic intermediates leading to hemifusion, followed by a single, rate-limiting step to pore formation. We interpret the series of intermediates preceding hemifusion as a result of the requirement that multiple copies of the trimeric hemagglutinin fusion protein be activated to initiate the fusion process.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Chernomordik LV, Kozlov MM. Protein-lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem. 2003;72:175–207. - PubMed
-
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. - PubMed
-
- Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

