Quantitative proteomic analysis of primary neurons reveals diverse changes in synaptic protein content in fmr1 knockout mice
- PMID: 18829439
- PMCID: PMC2563066
- DOI: 10.1073/pnas.0804678105
Quantitative proteomic analysis of primary neurons reveals diverse changes in synaptic protein content in fmr1 knockout mice
Abstract
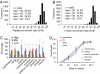
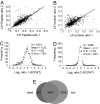
Fragile X syndrome (FXS) is a common inherited form of mental retardation that is caused, in the vast majority of cases, by the transcriptional silencing of a single gene, fmr1. The encoded protein, FMRP, regulates mRNA translation in neuronal dendrites, and it is thought that changes in translation-dependent forms of synaptic plasticity lead to many symptoms of FXS. However, little is known about the potentially extensive changes in synaptic protein content that accompany loss of FMRP. Here, we describe the development of a high-throughput quantitative proteomic method to identify differences in synaptic protein expression between wild-type and fmr1-/- mouse cortical neurons. The method is based on stable isotope labeling by amino acids in cell culture (SILAC), which has been used to characterize differentially expressed proteins in dividing cells, but not in terminally differentiated cells because of reduced labeling efficiency. To address the issue of incomplete labeling, we developed a mathematical method to normalize protein ratios relative to a reference based on the labeling efficiency. Using this approach, in conjunction with multidimensional protein identification technology (MudPIT), we identified >100 proteins that are up- or down-regulated. These proteins fall into a variety of functional categories, including those regulating synaptic structure, neurotransmission, dendritic mRNA transport, and several proteins implicated in epilepsy and autism, two endophenotypes of FXS. These studies provide insights into the potential origins of synaptic abnormalities in FXS and a demonstration of a methodology that can be used to explore neuronal protein changes in neurological disorders.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- de la Cruz FF. Fragile X syndrome. Am J Ment Defic. 1985;90:119–123. - PubMed
-
- Hagerman RJ. Fragile X-associated tremor/ataxia syndrome–an older face of the fragile X gene. J Dev Behav Pediatr. 2006;27:63–74. - PubMed
-
- Di Bonaventura C, et al. Status epilepticus in a patient with fragile X syndrome: Electro-clinical features and peri-ictal neuroimaging. Epileptic Disord. 2006;8:195–199. - PubMed
-
- Irwin SA, Galvez R, Greenough WT. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex. 2000;10:1038–1044. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

