Statistical challenges in preprocessing in microarray experiments in cancer
- PMID: 18829474
- PMCID: PMC3529914
- DOI: 10.1158/1078-0432.CCR-07-4532
Statistical challenges in preprocessing in microarray experiments in cancer
Abstract
Many clinical studies incorporate genomic experiments to investigate the potential associations between high-dimensional molecular data and clinical outcome. A critical first step in the statistical analyses of these experiments is that the molecular data are preprocessed. This article provides an overview of preprocessing methods, including summary algorithms and quality control metrics for microarrays. Some of the ramifications and effects that preprocessing methods have on the statistical results are illustrated. The discussions are centered around a microarray experiment based on lung cancer tumor samples with survival as the clinical outcome of interest. The procedures that are presented focus on the array platform used in this study. However, many of these issues are more general and are applicable to other instruments for genome-wide investigation. The discussions here will provide insight into the statistical challenges in preprocessing microarrays used in clinical studies of cancer. These challenges should not be viewed as inconsequential nuisances but rather as important issues that need to be addressed so that informed conclusions can be drawn.
Figures
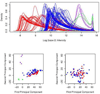
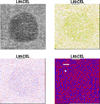
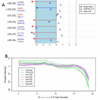
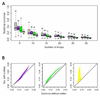
References
-
- Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nature Genetics. 1999;23(1):41–46. - PubMed
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene-expression patterns with a complementary-DNA microarray. Science. 1995;270(5235):467–470. - PubMed
-
- Barry WT, Nobel AB, Wright FA. Significance analysis of functional categories in gene expression studies: a structured permutation approach. Bioinformatics. 2005;21(9):1943–1949. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

