Deletions of CDKN2C in multiple myeloma: biological and clinical implications
- PMID: 18829482
- PMCID: PMC2581792
- DOI: 10.1158/1078-0432.CCR-08-0347
Deletions of CDKN2C in multiple myeloma: biological and clinical implications
Abstract
Purpose: Deletions of chromosome 1 have been described in 7% to 40% of cases of myeloma with inconsistent clinical consequences. CDKN2C at 1p32.3 has been identified in myeloma cell lines as the potential target of the deletion. We tested the clinical impact of 1p deletion and used high-resolution techniques to define the role of CDKN2C in primary patient material.
Experimental design: We analyzed 515 cases of monoclonal gammopathy of undetermined significance (MGUS), smoldering multiple myeloma (SMM), and newly diagnosed multiple myeloma using fluorescence in situ hybridization (FISH) for deletions of CDKN2C. In 78 myeloma cases, we carried out Affymetrix single nucleotide polymorphism mapping and U133 Plus 2.0 expression arrays. In addition, we did mutation, methylation, and Western blotting analysis.
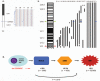
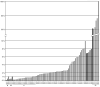
Results: By FISH we identified deletion of 1p32.3 (CDKN2C) in 3 of 66 MGUS (4.5%), 4 of 39 SMM (10.3%), and 55 of 369 multiple myeloma cases (15%). We examined the impact of copy number change at CDKN2C on overall survival (OS), and found that the cases with either hemizygous or homozygous deletion of CDKN2C had a worse OS compared with cases that were intact at this region (22 months versus 38 months; P = 0.003). Using gene mapping we identified three homozygous deletions at 1p32.3, containing CDKN2C, all of which lacked expression of CDKN2C. Cases with homozygous deletions of CDKN2C were the most proliferative myelomas, defined by an expression-based proliferation index, consistent with its biological function as a cyclin-dependent kinase inhibitor.
Conclusions: Our results suggest that deletions of CDKN2C are important in the progression and clinical outcome of myeloma.
Figures


References
-
- Morse L, Chen D, Franklin D, Xiong Y, Chen-Kiang S. Induction of cell cycle arrest and B cell terminal differentiation by CDK inhibitor p18(INK4c) and IL-6. Immunity. 1997;6:47–56. - PubMed
-
- Avet-Loiseau H, Andree-Ashley LE, Moore D, 2nd, et al. Molecular cytogenetic abnormalities in multiple myeloma and plasma cell leukemia measured using comparative genomic hybridization. Genes Chromosomes Cancer. 1997;19:124–33. - PubMed
-
- Cigudosa JC, Rao PH, Calasanz MJ, et al. Characterization of nonrandom chromosomal gains and losses in multiple myeloma by comparative genomic hybridization. Blood. 1998;91:3007–10. - PubMed
-
- Nilsson T, Hoglund M, Lenhoff S, et al. A pooled analysis of karyotypic patterns, breakpoints and imbalances in 783 cytogenetically abnormal multiple myelomas reveals frequently involved chromosome segments as well as significant age- and sex-related differences. Br J Haematol. 2003;120:960–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

