Salivary proteomics for oral cancer biomarker discovery
- PMID: 18829504
- PMCID: PMC2877125
- DOI: 10.1158/1078-0432.CCR-07-5037
Salivary proteomics for oral cancer biomarker discovery
Abstract
Purpose: This study aims to explore the presence of informative protein biomarkers in the human saliva proteome and to evaluate their potential for detection of oral squamous cell carcinoma (OSCC).
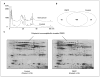
Experimental design: Whole saliva samples were collected from patients (n = 64) with OSCC and matched healthy subjects (n = 64). The proteins in pooled whole saliva samples of patients with OSCC (n = 16) and matched healthy subjects (n = 16) were profiled using shotgun proteomics based on C4 reversed-phase liquid chromatography for prefractionation, capillary reversed-phase liquid chromatography with quadruple time-of-flight mass spectrometry, and Mascot sequence database searching. Immunoassays were used for validation of the candidate biomarkers on a new group of OSCC (n = 48) and matched healthy subjects (n = 48). Receiver operating characteristic analysis was exploited to evaluate the diagnostic value of discovered candidate biomarkers for OSCC.
Results: Subtractive proteomics revealed several salivary proteins at differential levels between the OSCC patients and matched control subjects. Five candidate biomarkers were successfully validated using immunoassays on an independent set of OSCC patients and matched healthy subjects. The combination of these candidate biomarkers yielded a receiver operating characteristic value of 93%, sensitivity of 90%, and specificity of 83% in detecting OSCC.
Conclusion: Patient-based saliva proteomics is a promising approach to searching for OSCC biomarkers. The discovery of these new targets may lead to a simple clinical tool for the noninvasive diagnosis of oral cancer. Long-term longitudinal studies with large populations of individuals with oral cancer and those who are at high risk of developing oral cancer are needed to validate these potential biomarkers.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures

References
-
- Greenlee RT, Murray T, Bolden S, et al. Cancer tatistics, 2000. CA Cancer J Clin. 2000;50:7–33. - PubMed
-
- Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. - PubMed
-
- Cancer facts and figures 2006. Atlanta: American Cancer Society; 2006.
-
- Lippman SM, Hong WK. Molecular markers of the risk of oral cancer. N Engl J Med. 2001;344:1323–6. - PubMed
-
- Vokes EE, Weichselbaum RR, Lippman SM, et al. Head and neck cancer. N Engl J Med. 1993;328:184–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

