ATM and the Mre11-Rad50-Nbs1 complex respond to nucleoside analogue-induced stalled replication forks and contribute to drug resistance
- PMID: 18829552
- PMCID: PMC2631429
- DOI: 10.1158/0008-5472.CAN-08-0971
ATM and the Mre11-Rad50-Nbs1 complex respond to nucleoside analogue-induced stalled replication forks and contribute to drug resistance
Abstract
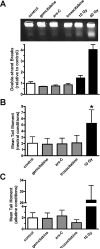
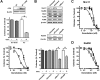
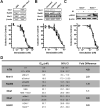
The Mre11-Rad50-Nbs1 complex and autophosphorylated Ser(1981)-ATM are involved in recognizing and repairing DNA damage, such as double-strand breaks (DSB). However, the role of these factors in response to stalled replication forks is not clear. Nucleoside analogues are agents that are incorporated into DNA during replication, which cause stalling of replication forks. The molecular mechanisms that sense these events may signal for DNA repair and contribute to survival but are poorly understood. Cellular responses to both DSBs and stalled replication forks are marked by H2AX phosphorylation on Ser(139) (gamma-H2AX), which forms nuclear foci at sites of DNA damage. Here, concentrations of the nucleoside analogues 1-beta-d-arabinofuranosylcytosine (cytarabine; ara-C), gemcitabine, and troxacitabine, which inhibited DNA synthesis by 90% within 2 hours, were determined for each agent. Using gamma-H2AX as a marker for changes in chromatin structure, we show that Mre11, Rad50, Nbs1, and phosphorylated ATM respond to nucleoside analogue-induced stalled replication forks by forming nuclear foci that colocalize with gamma-H2AX within 2 hours. Because neither DSBs nor single-strand breaks were detectable after nucleoside analogue exposure, we conclude that this molecular response is not due to the presence of DNA breaks. Deficiencies in ATM, Mre11, or Rad50 led to a 2- to 5-fold increase in clonogenic sensitization to gemcitabine, whereas Nbs1 and H2AX deficiency did not affect reproductive growth. Taken together, these results suggest that ATM, Mre11, and Rad50 are required for survival after replication fork stalling, whereas Nbs1 and H2AX are inconsequential.
Figures


References
-
- Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 2007;6:953–66. - PubMed
-
- Sampath D, Rao VA, Plunkett W. Mechanisms of apoptosis induction by nucleoside analogs. Oncogene. 2003;22:9063–74. - PubMed
-
- Sampath D, Shi Z, Plunkett W. Inhibition of cyclin-dependent kinase 2 by the Chk1-Cdc25A pathway during the S-phase checkpoint activated by fludarabine: dysregulation by 7-hydroxystaurosporine. Mol Pharmacol. 2002;62:680–8. - PubMed
-
- Ewald B, Sampath D, Plunkett W. H2AX phosphorylation marks gemcitabine-induced stalled replication forks and their collapse upon S-phase checkpoint abrogation. Mol Cancer Ther. 2007;6:1239–48. - PubMed
-
- Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci. 2006;31:402–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

