Identification of a residue in hepatitis C virus E2 glycoprotein that determines scavenger receptor BI and CD81 receptor dependency and sensitivity to neutralizing antibodies
- PMID: 18829747
- PMCID: PMC2593310
- DOI: 10.1128/JVI.01569-08
Identification of a residue in hepatitis C virus E2 glycoprotein that determines scavenger receptor BI and CD81 receptor dependency and sensitivity to neutralizing antibodies
Abstract
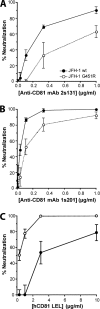
Hepatitis C virus (HCV) infection is dependent on at least three coreceptors: CD81, scavenger receptor BI (SR-BI), and claudin-1. The mechanism of how these molecules coordinate HCV entry is unknown. In this study we demonstrate that a cell culture-adapted JFH-1 mutant, with an amino acid change in E2 at position 451 (G451R), has a reduced dependency on SR-BI. This altered receptor dependency is accompanied by an increased sensitivity to neutralization by soluble CD81 and enhanced binding of recombinant E2 to cell surface-expressed and soluble CD81. Fractionation of HCV by density gradient centrifugation allows the analysis of particle-lipoprotein associations. The cell culture-adapted mutation alters the relationship between particle density and infectivity, with the peak infectivity occurring at higher density than the parental virus. No association was observed between particle density and SR-BI or CD81 coreceptor dependence. JFH-1 G451R is highly sensitive to neutralization by gp-specific antibodies, suggesting increased epitope exposure at the virion surface. Finally, an association was observed between JFH-1 particle density and sensitivity to neutralizing antibodies (NAbs), suggesting that lipoprotein association reduces the sensitivity of particles to NAbs. In summary, mutation of E2 at position 451 alters the relationship between particle density and infectivity, disrupts coreceptor dependence, and increases virion sensitivity to receptor mimics and NAbs. Our data suggest that a balanced interplay between HCV particles, lipoprotein components, and viral receptors allows the evasion of host immune responses.
Figures
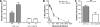



References
-
- Alberti, A., L. Chemello, and L. Benvegnu. 1999. Natural history of hepatitis C. J. Hepatol. 31(Suppl. 1)17-24. - PubMed
-
- Andreo, U., P. Maillard, O. Kalinina, M. Walic, E. Meurs, M. Martinot, P. Marcellin, and A. Budkowska. 2007. Lipoprotein lipase mediates hepatitis C virus (HCV) cell entry and inhibits HCV infection. Cell Microbiol. 92445-2456. - PubMed
-
- Barth, H., C. Schafer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. Van Kuppevelt, E. Depla, F. Von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 27841003-41012. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

