The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators
- PMID: 18829985
- PMCID: PMC2593676
- DOI: 10.1104/pp.108.124784
The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators
Abstract
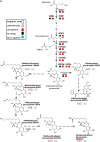
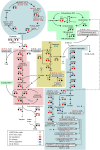
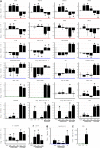
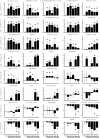
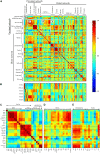
In this study, transcriptomics and metabolomics data were integrated in order to examine the regulation of glucosinolate (GS) biosynthesis in Arabidopsis (Arabidopsis thaliana) and its interface with pathways of primary metabolism. Our genetic material for analyses were transgenic plants overexpressing members of two clades of genes (ALTERED TRYPTOPHAN REGULATION1 [ATR1]-like and MYB28-like) that regulate the aliphatic and indole GS biosynthetic pathways (AGs and IGs, respectively). We show that activity of these regulators is not restricted to the metabolic space surrounding GS biosynthesis but is tightly linked to more distal metabolic networks of primary metabolism. This suggests that with similarity to the regulators we have investigated here, other factors controlling pathways of secondary metabolism might also control core pathways of central metabolism. The relatively broad view of transcripts and metabolites altered in transgenic plants overexpressing the different factors underlined novel links of GS metabolism to additional metabolic pathways, including those of jasmonic acid, folate, benzoic acid, and various phenylpropanoids. It also revealed transcriptional and metabolic hubs in the "distal" network of metabolic pathways supplying precursors to GS biosynthesis and that overexpression of the ATR1-like clade genes has a much broader effect on the metabolism of indolic compounds than described previously. While the reciprocal, negative cross talk between the methionine and tryptophan pathways that generate GSs in Arabidopsis has been suggested previously, we now show that it is not restricted to AGs and IGs but includes additional metabolites, such as the phytoalexin camalexin. Combining the profiling data of transgenic lines with gene expression correlation analysis allowed us to propose a model of how the balance in the metabolic network is maintained by the GS biosynthesis regulators. It appears that ATR1/MYB34 is an important mediator between the gene activities of the two clades. While it is very similar to the ATR1-like clade members in terms of downstream gene targets, its expression is highly correlated with that of the MYB28-like clade members. Finally, we used the unique transgenic plants obtained here to show that AGs are likely more potent deterrents of the whitefly Bemisia tabaci compared with IGs. The influence on insect behavior raises an important question for future investigation of the functional aspect of our initial finding, which pointed to enriched expression of the MYB28-like clade genes in the abaxial domain of the Arabidopsis leaf.
Figures


Similar articles
-
The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana.Plant J. 2007 Jul;51(2):247-61. doi: 10.1111/j.1365-313X.2007.03133.x. Epub 2007 May 23. Plant J. 2007. PMID: 17521412
-
Novel insights into the function of Arabidopsis R2R3-MYB transcription factors regulating aliphatic glucosinolate biosynthesis.Plant Cell Physiol. 2013 Aug;54(8):1335-44. doi: 10.1093/pcp/pct085. Epub 2013 Jun 20. Plant Cell Physiol. 2013. PMID: 23792303
-
The Glucosinolate Biosynthetic Gene AOP2 Mediates Feed-back Regulation of Jasmonic Acid Signaling in Arabidopsis.Mol Plant. 2015 Aug;8(8):1201-12. doi: 10.1016/j.molp.2015.03.001. Epub 2015 Mar 7. Mol Plant. 2015. PMID: 25758208
-
A Comprehensive Gene Inventory for Glucosinolate Biosynthetic Pathway in Arabidopsis thaliana.J Agric Food Chem. 2020 Jul 15;68(28):7281-7297. doi: 10.1021/acs.jafc.0c01916. Epub 2020 Jul 1. J Agric Food Chem. 2020. PMID: 32551569 Review.
-
Regulation of glucosinolate biosynthesis.J Exp Bot. 2021 Jan 20;72(1):70-91. doi: 10.1093/jxb/eraa479. J Exp Bot. 2021. PMID: 33313802 Review.
Cited by
-
Genetic analysis of glucosinolate variability in broccoli florets using genome-anchored single nucleotide polymorphisms.Theor Appl Genet. 2015 Jul;128(7):1431-47. doi: 10.1007/s00122-015-2517-x. Epub 2015 May 1. Theor Appl Genet. 2015. PMID: 25930056
-
The impact of the absence of aliphatic glucosinolates on water transport under salt stress in Arabidopsis thaliana.Front Plant Sci. 2015 Jul 15;6:524. doi: 10.3389/fpls.2015.00524. eCollection 2015. Front Plant Sci. 2015. PMID: 26236322 Free PMC article.
-
Tilting Plant Metabolism for Improved Metabolite Biosynthesis and Enhanced Human Benefit.Molecules. 2015 Jul 13;20(7):12698-731. doi: 10.3390/molecules200712698. Molecules. 2015. PMID: 26184148 Free PMC article. Review.
-
The effects of glucosinolates and their breakdown products on necrotrophic fungi.PLoS One. 2013 Aug 5;8(8):e70771. doi: 10.1371/journal.pone.0070771. Print 2013. PLoS One. 2013. PMID: 23940639 Free PMC article.
-
Diurnal and light regulation of sulphur assimilation and glucosinolate biosynthesis in Arabidopsis.J Exp Bot. 2013 Feb;64(4):1039-48. doi: 10.1093/jxb/ers378. Epub 2013 Jan 10. J Exp Bot. 2013. PMID: 23314821 Free PMC article.
References
-
- Aloni R, Schwalm K, Langhans M, Ullrich CI (2003) Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216 841–853 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 - PubMed
-
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 7 289–300
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

