Evolution of the aging brain transcriptome and synaptic regulation
- PMID: 18830410
- PMCID: PMC2553198
- DOI: 10.1371/journal.pone.0003329
Evolution of the aging brain transcriptome and synaptic regulation
Abstract
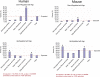
Alzheimer's disease and other neurodegenerative disorders of aging are characterized by clinical and pathological features that are relatively specific to humans. To obtain greater insight into how brain aging has evolved, we compared age-related gene expression changes in the cortex of humans, rhesus macaques, and mice on a genome-wide scale. A small subset of gene expression changes are conserved in all three species, including robust age-dependent upregulation of the neuroprotective gene apolipoprotein D (APOD) and downregulation of the synaptic cAMP signaling gene calcium/calmodulin-dependent protein kinase IV (CAMK4). However, analysis of gene ontology and cell type localization shows that humans and rhesus macaques have diverged from mice due to a dramatic increase in age-dependent repression of neuronal genes. Many of these age-regulated neuronal genes are associated with synaptic function. Notably, genes associated with GABA-ergic inhibitory function are robustly age-downregulated in humans but not in mice at the level of both mRNA and protein. Gene downregulation was not associated with overall neuronal or synaptic loss. Thus, repression of neuronal gene expression is a prominent and recently evolved feature of brain aging in humans and rhesus macaques that may alter neural networks and contribute to age-related cognitive changes.
Conflict of interest statement
Figures





References
-
- Martin GM. Genetic modulation of senescent phenotypes in Homo sapiens. Cell. 2005;120:523–532. - PubMed
-
- Yankner BA, Lu T, Loerch P. The Aging Brain. Annu Rev Pathol. 2007;3:41–66. - PubMed
-
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. - PubMed
-
- Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

