Fatty acid synthase is up-regulated during hepatitis C virus infection and regulates hepatitis C virus entry and production
- PMID: 18830996
- PMCID: PMC2614928
- DOI: 10.1002/hep.22508
Fatty acid synthase is up-regulated during hepatitis C virus infection and regulates hepatitis C virus entry and production
Abstract
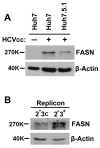
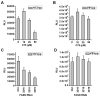
Hepatitis C virus (HCV) is a major human pathogen that causes serious illness, including acute and chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Using a mass spectrometry-based proteomics approach, we have identified 175 proteins from a cell culture supernatant fraction containing the HCV genotype 2a (JFH1) virus, among which fatty acid synthase (FASN), the multifunctional enzyme catalyzing the de novo synthesis of fatty acids, was confirmed to be highly enriched. Subsequent studies showed that FASN expression increased in the human hepatoma cell line, Huh7, or its derivative, upon HCV infection. Blocking FASN activity by C75, a pharmacological FASN inhibitor, led to decreased HCV production. Reduction of FASN by RNA interference suppressed viral replication in both replicon and infection systems. Remarkably, FASN appeared to be selectively required for the expression of claudin-1, a tight junction protein that was recently identified as an entry coreceptor for HCV, but not for the expression of another HCV coreceptor, CD81. The decrease in Claudin-1 expression resulting from FASN inhibition was accompanied by a decrease in transepithelial electric resistance of Huh7 cells, implying a reduction in the relative tightness of the cell monolayer. Consequently, the entry of human immunodeficiency virus-HCV pseudotypes was significantly inhibited in C75-treated Huh7 cells.
Conclusion: As far as we know, this is the first line of evidence that demonstrates that HCV infection directly induces FASN expression, and thus suggests a possible mechanism by which HCV infection alters the cellular lipid profile and causes diseases such as steatosis.
Conflict of interest statement
Figures




References
-
- Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. - PubMed
-
- Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. - PubMed
-
- Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. - PubMed
-
- Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39:5–19. - PubMed
-
- Bjornsson E, Angulo P. Hepatitis C and steatosis. Arch Med Res. 2007;38:621–627. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
