Covalent binding of flavins to RnfG and RnfD in the Rnf complex from Vibrio cholerae
- PMID: 18831535
- PMCID: PMC2643342
- DOI: 10.1021/bi800920j
Covalent binding of flavins to RnfG and RnfD in the Rnf complex from Vibrio cholerae
Abstract
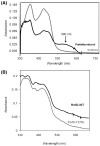
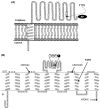
Enzymes of the Rnf family are believed to be bacterial redox-driven ion pumps, coupling an oxidoreduction process to the translocation of Na+ across the cell membrane. Here we show for the first time that Rnf is a flavoprotein, with FMN covalently bound to threonine-175 in RnfG and a second flavin bound to threonine-187 in RnfD. Rnf subunits D and G are homologous to subunits B and C of Na+-NQR, respectively. Each of these Na+-NQR subunits includes a conserved S(T)GAT motif, with FMN covalently bound to the final threonine. RnfD and RnfG both contain the same motif, suggesting that they bind flavins in a similar way. In order to investigate this, the genes for RnfD and RnfG from Vibrio cholerae were cloned and expressed individually in that organism. In both cases the produced protein fluoresced under UV illumination on an SDS gel, further indicating the presence of flavin. However, analysis of the mutants RnfG-T175L, RnfD-T278L, and RnfD-T187V showed that RnfG-T175 and RnfD-T187 are the likely flavin ligands. This indicates that, in the case of RnfD, the flavin is bound, not to the SGAT sequence but to the final residues of a TMAT sequence, a novel variant of the flavin binding motif. In the case of RnfG, flavin analysis, followed by MALDI-TOF-TOF mass spectrometry, showed that an FMN is covalently attached to threonine-175, the final threonine of the S(T)GAT sequence. Studies by visible, EPR, and ENDOR spectroscopy showed that, upon partial reduction, the isolated RnfG produces a neutral semiquinone intermediate. The semiquinone species disappeared upon full reduction and was not observed in the denatured protein. A topological analysis combining reporter protein fusion and computer predictions indicated that the flavins in RnfG and RnfD are localized in the periplasmic space. In contrast, in NqrC and NqrB the flavins are located in a cytoplasmic loop. This topological analysis suggests that there may be mechanistic differences between the Rnf and Na+-NQR complexes.
Figures




References
-
- Schmehl M, Jahn A, Meyer zu Vilsendorf A, Hennecke S, Masepohl B, Schuppler M, Marxer M, Oelze J, Klipp W. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet. 1993;241:602–615. - PubMed
-
- Jouanneau Y, Jeong HS, Hugo N, Meyer C, Willison JC. Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus-characterization of two membrane-bound iron-sulfur proteins. Eur. J. Biochem. 1998;251:54–64. - PubMed
-
- Kumagai H, Fujiwara T, Matsubara H, Saeki K. Membrane localization, topology, and mutual stabilization of the rnfABC gene products in Rhodobacter capsulatus and implications for a new family of energy-coupling NADH oxidoreductases. Biochemistry. 1997;36:5509–5521. - PubMed
-
- Duran-Pinedo AENK, Duncan MJ. The RprY response regulator of Porphyromonas gingivalis. Mol. Microbiol. 2007;64:1061–1074. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

