Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo
- PMID: 18832072
- PMCID: PMC2559904
- DOI: 10.1101/gad.486108
Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo
Abstract
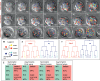
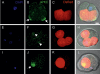
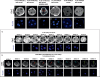
Genesis of the trophectoderm and inner cell mass (ICM) lineages occurs in two stages. It is initiated via asymmetric divisions of eight- and 16-cell blastomeres that allocate cells to inner and outer positions, each with different developmental fates. Outside cells become committed to the trophectoderm at the blastocyst stage through Cdx2 activity, but here we show that Cdx2 can also act earlier to influence cell allocation. Increasing Cdx2 levels in individual blastomeres promotes symmetric divisions, thereby allocating more cells to the trophectoderm, whereas reducing Cdx2 promotes asymmetric divisions and consequently contribution to the ICM. Furthermore, both Cdx2 mRNA and protein levels are heterogeneous at the eight-cell stage. This heterogeneity depends on cell origin and has developmental consequences. Cdx2 expression is minimal in cells with unrestricted developmental potential that contribute preferentially to the ICM and is maximal in cells with reduced potential that contribute more to the trophectoderm. Finally, we describe a mutually reinforcing relationship between cellular polarity and Cdx2: Cdx2 influences cell polarity by up-regulating aPKC, but cell polarity also influences Cdx2 through asymmetric distribution of Cdx2 mRNA in polarized blastomeres. Thus, divisions generating inside and outside cells are truly asymmetric with respect to cell fate instructions. These two interacting effects ensure the generation of a stable outer epithelium by the blastocyst stage.
Figures




References
-
- Chazaud C., Yamanaka Y., Pawson T., Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2–MAPK pathway. Dev. Cell. 2006;10:615–624. - PubMed
-
- Ciruna B., Rossant J. FGF signalling regulates mesoderm cell fate specification and morphogenetic movement at the primitive steak. Dev. Cell. 2001;1:37–49. - PubMed
-
- Dietrich J.E., Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
