Sequestration of Polo kinase to microtubules by phosphopriming-independent binding to Map205 is relieved by phosphorylation at a CDK site in mitosis
- PMID: 18832073
- PMCID: PMC2559908
- DOI: 10.1101/gad.486808
Sequestration of Polo kinase to microtubules by phosphopriming-independent binding to Map205 is relieved by phosphorylation at a CDK site in mitosis
Abstract
The conserved Polo kinase controls multiple events in mitosis and cytokinesis. Although Polo-like kinases are regulated by phosphorylation and proteolysis, control of subcellular localization plays a major role in coordinating their mitotic functions. This is achieved largely by the Polo-Box Domain, which binds prephosphorylated targets. However, it remains unclear whether and how Polo might interact with partner proteins when priming mitotic kinases are inactive. Here we show that Polo associates with microtubules in interphase and cytokinesis, through a strong interaction with the microtubule-associated protein Map205. Surprisingly, this interaction does not require priming phosphorylation of Map205, and the Polo-Box Domain of Polo is required but not sufficient for this interaction. Moreover, phosphorylation of Map205 at a CDK site relieves this interaction. Map205 can stabilize Polo and inhibit its cellular activity in vivo. In syncytial embryos, the centrosome defects observed in polo hypomorphs are enhanced by overexpression of Map205 and suppressed by its deletion. We propose that Map205-dependent targeting of Polo to microtubules provides a stable reservoir of Polo that can be rapidly mobilized by the activity of Cdk1 at mitotic entry.
Figures
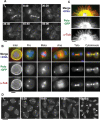

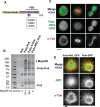


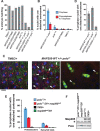
References
-
- Ahonen L.J., Kallio M.J., Daum J.R., Bolton M., Manke I.A., Yaffe M.B., Stukenberg P.T., Gorbsky G.J. Polo-like kinase 1 creates the tension-sensing 3F3/2 phosphoepitope and modulates the association of spindle-checkpoint proteins at kinetochores. Curr. Biol. 2005;15:1078–1089. - PubMed
-
- Alexandru G., Uhlmann F., Mechtler K., Poupart M.A., Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. - PubMed
-
- Andersen S.S. Spindle assembly and the art of regulating microtubule dynamics by MAPs and Stathmin/Op18. Trends Cell Biol. 2000;10:261–267. - PubMed
-
- Barr F.A., Sillje H.H., Nigg E.A. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004;5:429–440. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
