Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae
- PMID: 18832470
- PMCID: PMC2572952
- DOI: 10.1073/pnas.0803654105
Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae
Abstract
Bacterial populations are subject to complex processes of diversification that involve mutation and horizontal DNA transfer mediated by transformation, transduction, or conjugation. Tracing the evolutionary events leading to genetic changes allows us to infer the history of a microbe. Here, we combine experimental and in silico approaches to explore the forces that drive the genome dynamics of Streptococcus agalactiae, the leading cause of neonatal infections. We demonstrate that large DNA segments of up to 334 kb of the chromosome of S. agalactiae can be transferred through conjugation from multiple initiation sites. Consistently, a genome-wide map analysis of nucleotide polymorphisms among eight human isolates demonstrated that each chromosome is a mosaic of large chromosomal fragments from different ancestors suggesting that large DNA exchanges have contributed to the genome dynamics in the natural population. The analysis of the resulting genetic flux led us to propose a model for the evolutionary history of this species in which clonal complexes of clinical importance derived from a single clone that evolved by exchanging large chromosomal regions with more distantly related strains. The emergence of this clone could be linked to selective sweeps associated with the reduction of genetic diversity in three regions within a large panel of human isolates. Up to now sex in bacteria has been assumed to involve mainly small regions; our results define S. agalactiae as an alternative paradigm in the study of bacterial evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
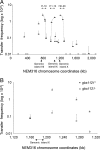

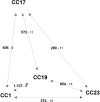

References
-
- Smith JM, Dowson CG, Spratt BG. Localized sex in bacteria. Nature. 1991;349:29–31. - PubMed
-
- Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–687. - PubMed
-
- Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–721. - PubMed
-
- Spratt BG. Exploring the concept of clonality in bacteria. Methods Mol Biol. 2004;266:323–352. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

