Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis
- PMID: 18832610
- PMCID: PMC2698935
- DOI: 10.1126/science.1163885
Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis
Abstract
The protein modifier ubiquitin is a signal for proteasome-mediated degradation in eukaryotes. Proteasome-bearing prokaryotes have been thought to degrade proteins via a ubiquitin-independent pathway. We have identified a prokaryotic ubiquitin-like protein, Pup (Rv2111c), which was specifically conjugated to proteasome substrates in the pathogen Mycobacterium tuberculosis. Pupylation occurred on lysines and required proteasome accessory factor A (PafA). In a pafA mutant, pupylated proteins were absent and substrates accumulated, thereby connecting pupylation with degradation. Although analogous to ubiquitylation, pupylation appears to proceed by a different chemistry. Thus, like eukaryotes, bacteria may use a small-protein modifier to control protein stability.
Figures


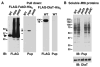

Comment in
-
Microbiology. A protein pupylation paradigm.Science. 2008 Nov 14;322(5904):1062-3. doi: 10.1126/science.1166485. Science. 2008. PMID: 19008436 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

