Aberrant DNA methylation is a dominant mechanism in MDS progression to AML
- PMID: 18832655
- PMCID: PMC2637194
- DOI: 10.1182/blood-2008-06-163246
Aberrant DNA methylation is a dominant mechanism in MDS progression to AML
Abstract
Myelodysplastic syndromes (MDSs) are clonal hematologic disorders that frequently represent an intermediate disease stage before progression to acute myeloid leukemia (AML). As such, study of MDS/AML can provide insight into the mechanisms of neoplastic evolution. In 184 patients with MDS and AML, DNA methylation microarray and high-density single nucleotide polymorphism array (SNP-A) karyotyping were used to assess the relative contributions of aberrant DNA methylation and chromosomal deletions to tumor-suppressor gene (TSG) silencing during disease progression. Aberrant methylation was seen in every sample, on average affecting 91 of 1505 CpG loci in early MDS and 179 of 1505 loci after blast transformation (refractory anemia with excess blasts [RAEB]/AML). In contrast, chromosome aberrations were seen in 79% of early MDS samples and 90% of RAEB/AML samples, and were not as widely distributed over the genome. Analysis of the most frequently aberrantly methylated genes identified FZD9 as a candidate TSG on chromosome 7. In patients with chromosome deletion at the FZD9 locus, aberrant methylation of the remaining allele was associated with the poorest clinical outcome. These results indicate that aberrant methylation can cooperate with chromosome deletions to silence TSG. However, the ubiquity, extent, and correlation with disease progression suggest that aberrant DNA methylation is the dominant mechanism for TSG silencing and clonal variation in MDS evolution to AML.
Figures
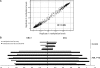
 indicates the array average methylation values (β-value) for the same CpG sites.
indicates the array average methylation values (β-value) for the same CpG sites.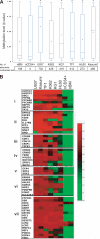



 represents the degree of FZD9 methylation in the sample; ■, the FZD9 expression levels determined by semiquantitative RT-PCR, defined as fold change compared with the control marked with an asterisk. (D) FZD9 CpG methylation is not part of a wider hypermethylation at its chromosome 7 locus. The height of the vertical bars represents the frequency of aberrant hypermethylation of the CpG sites designated along the horizontal line. The space between bars marked with 0 indicates analyzed CpG sites that were not hypermethylated in any patient. (E) Regions of chromosome deletion and UPD, detected by SNP-A, that involves the FZD9 locus. FZD9 is designated by the vertical red bar. Red lines depict single SNP signal intensity, whereas green lines present an average value of SNP signal intensity. The horizontal purple bar represents areas of chromosome deletion, whereas the horizontal blue bar represents areas of loss of heterozygosity through UPD. (F) Chromosomal deletion or duplication of an FZD9 allele, combined with aberrant methylation of the remaining allele, is associated with a worse prognosis than either abnormality alone.
represents the degree of FZD9 methylation in the sample; ■, the FZD9 expression levels determined by semiquantitative RT-PCR, defined as fold change compared with the control marked with an asterisk. (D) FZD9 CpG methylation is not part of a wider hypermethylation at its chromosome 7 locus. The height of the vertical bars represents the frequency of aberrant hypermethylation of the CpG sites designated along the horizontal line. The space between bars marked with 0 indicates analyzed CpG sites that were not hypermethylated in any patient. (E) Regions of chromosome deletion and UPD, detected by SNP-A, that involves the FZD9 locus. FZD9 is designated by the vertical red bar. Red lines depict single SNP signal intensity, whereas green lines present an average value of SNP signal intensity. The horizontal purple bar represents areas of chromosome deletion, whereas the horizontal blue bar represents areas of loss of heterozygosity through UPD. (F) Chromosomal deletion or duplication of an FZD9 allele, combined with aberrant methylation of the remaining allele, is associated with a worse prognosis than either abnormality alone.References
-
- Woods WG, Nesbit ME, Buckley J, et al. Correlation of chromosome abnormalities with patient characteristics, histologic subtype, and induction success in children with acute nonlymphocytic leukemia. J Clin Oncol. 1985;3:3–11. - PubMed
-
- Yunis JJ, Rydell RE, Oken MM, et al. Refined chromosome analysis as an independent prognostic indicator in de novo myelodysplastic syndromes. Blood. 1986;67:1721–1730. - PubMed
-
- Billstrom R, Thiede T, Hansen S, et al. Bone marrow karyotype and prognosis in primary myelodysplastic syndromes. Eur J Haematol. 1988;41:341–346. - PubMed
-
- Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2:S4–S11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

