An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies
- PMID: 18833193
- PMCID: PMC2572182
- DOI: 10.1038/emboj.2008.201
An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies
Abstract
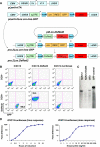
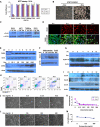
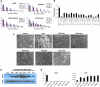
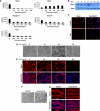
Facioscapulohumeral muscular dystrophy (FSHD) is caused by an unusual deletion with neomorphic activity. This deletion derepresses genes in cis; however which candidate gene causes the FSHD phenotype, and through what mechanism, is unknown. We describe a novel genetic tool, inducible cassette exchange, enabling rapid generation of isogenetically modified cells with conditional and variable transgene expression. We compare the effects of expressing variable levels of each FSHD candidate gene on myoblasts. This screen identified only one gene with overt toxicity: DUX4 (double homeobox, chromosome 4), a protein with two homeodomains, each similar in sequence to Pax3 and Pax7. DUX4 expression recapitulates key features of the FSHD molecular phenotype, including repression of MyoD and its target genes, diminished myogenic differentiation, repression of glutathione redox pathway components, and sensitivity to oxidative stress. We further demonstrate competition between DUX4 and Pax3/Pax7: when either Pax3 or Pax7 is expressed at high levels, DUX4 is no longer toxic. We propose a hypothesis for FSHD in which DUX4 expression interferes with Pax7 in satellite cells, and inappropriately regulates Pax targets, including myogenic regulatory factors, during regeneration.
Figures

References
-
- Agha-Mohammadi S, O'Malley M, Etemad A, Wang Z, Xiao X, Lotze MT (2004) Second-generation tetracycline-regulatable promoter: repositioned tet operator elements optimize transactivator synergy while shorter minimal promoter offers tight basal leakiness. J Gene Med 6: 817–828 - PubMed
-
- Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA (2007) Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell 130: 349–362 - PubMed
-
- Buckingham M, Relaix F (2007) The role of pax genes in the development of tissues and organs: pax3 and pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol 23: 645–673 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

