Curing of the [URE3] prion by Btn2p, a Batten disease-related protein
- PMID: 18833194
- PMCID: PMC2572181
- DOI: 10.1038/emboj.2008.198
Curing of the [URE3] prion by Btn2p, a Batten disease-related protein
Abstract
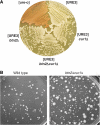
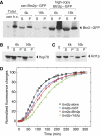
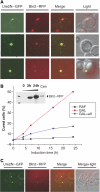
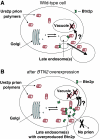
[URE3] is a prion (infectious protein), a self-propagating amyloid form of Ure2p, a regulator of yeast nitrogen catabolism. We find that overproduction of Btn2p, or its homologue Ypr158 (Cur1p), cures [URE3]. Btn2p is reported to be associated with late endosomes and to affect sorting of several proteins. We find that double deletion of BTN2 and CUR1 stabilizes [URE3] against curing by several agents, produces a remarkable increase in the proportion of strong [URE3] variants arising de novo and an increase in the number of [URE3] prion seeds. Thus, normal levels of Btn2p and Cur1p affect prion generation and propagation. Btn2p-green fluorescent protein (GFP) fusion proteins appear as a single dot located close to the nucleus and the vacuole. During the curing process, those cells having both Ure2p-GFP aggregates and Btn2p-RFP dots display striking colocalization. Btn2p curing requires cell division, and our results suggest that Btn2p is part of a system, reminiscent of the mammalian aggresome, that collects aggregates preventing their efficient distribution to progeny cells.
Figures




Similar articles
-
Normal levels of the antiprion proteins Btn2 and Cur1 cure most newly formed [URE3] prion variants.Proc Natl Acad Sci U S A. 2014 Jul 1;111(26):E2711-20. doi: 10.1073/pnas.1409582111. Epub 2014 Jun 17. Proc Natl Acad Sci U S A. 2014. PMID: 24938787 Free PMC article.
-
Innate immunity to yeast prions: Btn2p and Cur1p curing of the [URE3] prion is prevented by 60S ribosomal protein deficiency or ubiquitin/proteasome system overactivity.Genetics. 2021 Apr 15;217(4):iyab013. doi: 10.1093/genetics/iyab013. Genetics. 2021. PMID: 33857305 Free PMC article.
-
Proteasome Control of [URE3] Prion Propagation by Degradation of Anti-Prion Proteins Cur1 and Btn2 in Saccharomyces cerevisiae.Genetics. 2021 May 17;218(1):iyab037. doi: 10.1093/genetics/iyab037. Genetics. 2021. PMID: 33742650 Free PMC article.
-
Innate immunity to prions: anti-prion systems turn a tsunami of prions into a slow drip.Curr Genet. 2021 Dec;67(6):833-847. doi: 10.1007/s00294-021-01203-1. Epub 2021 Jul 28. Curr Genet. 2021. PMID: 34319422 Review.
-
Protein-based inheritance in Saccharomyces cerevisiae: [URE3] as a prion form of the nitrogen regulatory protein Ure2.Res Microbiol. 2001 Sep;152(7):605-12. doi: 10.1016/s0923-2508(01)01239-6. Res Microbiol. 2001. PMID: 11605980 Review.
Cited by
-
Metabolic and chaperone gene loss marks the origin of animals: evidence for Hsp104 and Hsp78 chaperones sharing mitochondrial enzymes as clients.PLoS One. 2015 Feb 24;10(2):e0117192. doi: 10.1371/journal.pone.0117192. eCollection 2015. PLoS One. 2015. PMID: 25710177 Free PMC article.
-
Sequestrase chaperones protect against oxidative stress-induced protein aggregation and [PSI+] prion formation.PLoS Genet. 2024 Feb 29;20(2):e1011194. doi: 10.1371/journal.pgen.1011194. eCollection 2024 Feb. PLoS Genet. 2024. PMID: 38422160 Free PMC article.
-
Overexpression of Hsp104 by Causing Dissolution of the Prion Seeds Cures the Yeast [PSI+] Prion.Int J Mol Sci. 2023 Jun 29;24(13):10833. doi: 10.3390/ijms241310833. Int J Mol Sci. 2023. PMID: 37446010 Free PMC article.
-
Prions in Microbes: The Least in the Most.J Microbiol. 2023 Oct;61(10):881-889. doi: 10.1007/s12275-023-00070-4. Epub 2023 Sep 5. J Microbiol. 2023. PMID: 37668956 Review.
-
Btn3 is a negative regulator of Btn2-mediated endosomal protein trafficking and prion curing in yeast.Mol Biol Cell. 2011 May 15;22(10):1648-63. doi: 10.1091/mbc.E10-11-0878. Epub 2011 Mar 25. Mol Biol Cell. 2011. PMID: 21441304 Free PMC article.
References
-
- Allen KD, Chernova TA, Tennant EP, Wilkinson KD, Chernoff YO (2007) Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J Biol Chem 282: 3004–3013 - PubMed
-
- Baxa U, Wickner RB, Steven AC, Anderson D, Marekov L, Yau W-M, Tycko R (2007) Characterization of β-sheet structure in Ure2p1-89 yeast prion fibrils by solid state nuclear magnetic resonance. Biochemistry 46: 13149–13162 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

