Sequential tyrosine sulfation of CXCR4 by tyrosylprotein sulfotransferases
- PMID: 18834145
- PMCID: PMC2662774
- DOI: 10.1021/bi800965m
Sequential tyrosine sulfation of CXCR4 by tyrosylprotein sulfotransferases
Abstract
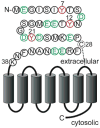
CXC-chemokine receptor 4 (CXCR4) is a G protein-coupled receptor for stromal cell-derived factor-1 (SDF-1/CXCL12). SDF-1-induced CXCR4 signaling is indispensable for embryonic development and crucial for immune cell homing and has been implicated in metastasis of numerous types of cancer. CXCR4 also serves as the major coreceptor for cellular entry of T-cell line-tropic (X4) HIV-1 strains. Tyrosine residues in the N-terminal tail of CXCR4, which are post-translationally sulfated, are implicated in the high-affinity binding of SDF-1 to CXCR4. However, the specific roles of three potential tyrosine sulfation sites are not well understood. We investigated the pattern and sequence of CXCR4 sulfation by using recombinant human tyrosylprotein sulfotransferases TPST-1 and TPST-2 to modify a peptide that corresponds to amino acids 1-38 of the receptor (CXCR4 1-38). We analyzed the reaction products with a combination of reversed-phase HPLC, proteolytic cleavage, and mass spectrometry. We found that CXCR4 1-38 is sulfated efficiently by both TPST enzymes, leading to a final product with three sulfotyrosine residues. Sulfates were added stepwise to the peptide, producing specific intermediates with one or two sulfotyrosines. The pattern of sulfation in these intermediates indicates that with both enzymes Tyr-21 is sulfated first, followed by Tyr-12 or Tyr-7. Using heteronuclear NMR spectroscopy, we demonstrated that the SDF-1 binding affinity of CXCR4 1-38 increases with the number of sulfotyrosines present, which suggests a potential physiological role for sulfation of all three sites in the N-terminus of CXCR4. These results provide a structural basis for understanding the role of post-translational tyrosine sulfation in SDF-1-induced CXCR4 signaling.
Figures


Similar articles
-
Tyrosine sulfation of CCR5 N-terminal peptide by tyrosylprotein sulfotransferases 1 and 2 follows a discrete pattern and temporal sequence.Proc Natl Acad Sci U S A. 2002 Aug 20;99(17):11031-6. doi: 10.1073/pnas.172380899. Epub 2002 Aug 8. Proc Natl Acad Sci U S A. 2002. PMID: 12169668 Free PMC article.
-
Preparation and Analysis of N-Terminal Chemokine Receptor Sulfopeptides Using Tyrosylprotein Sulfotransferase Enzymes.Methods Enzymol. 2016;570:357-88. doi: 10.1016/bs.mie.2015.09.004. Epub 2015 Nov 14. Methods Enzymol. 2016. PMID: 26921955 Free PMC article.
-
Recognition of a CXCR4 sulfotyrosine by the chemokine stromal cell-derived factor-1alpha (SDF-1alpha/CXCL12).J Mol Biol. 2006 Jun 23;359(5):1400-9. doi: 10.1016/j.jmb.2006.04.052. Epub 2006 May 11. J Mol Biol. 2006. PMID: 16725153 Free PMC article.
-
Tyrosine sulfation: an increasingly recognised post-translational modification of secreted proteins.N Biotechnol. 2009 Jun;25(5):299-317. doi: 10.1016/j.nbt.2009.03.011. N Biotechnol. 2009. PMID: 19658209 Review.
-
Determinants of tyrosylprotein sulfation coding and substrate specificity of tyrosylprotein sulfotransferases in metazoans.Chem Biol Interact. 2016 Nov 25;259(Pt A):17-22. doi: 10.1016/j.cbi.2016.04.006. Epub 2016 Apr 7. Chem Biol Interact. 2016. PMID: 27062897 Review.
Cited by
-
Tyrosine sulfation of chemokine receptor CCR2 enhances interactions with both monomeric and dimeric forms of the chemokine monocyte chemoattractant protein-1 (MCP-1).J Biol Chem. 2013 Apr 5;288(14):10024-10034. doi: 10.1074/jbc.M112.447359. Epub 2013 Feb 13. J Biol Chem. 2013. PMID: 23408426 Free PMC article.
-
Structural basis for the broad substrate specificity of the human tyrosylprotein sulfotransferase-1.Sci Rep. 2017 Aug 18;7(1):8776. doi: 10.1038/s41598-017-07141-8. Sci Rep. 2017. PMID: 28821720 Free PMC article.
-
Heparin oligosaccharides inhibit chemokine (CXC motif) ligand 12 (CXCL12) cardioprotection by binding orthogonal to the dimerization interface, promoting oligomerization, and competing with the chemokine (CXC motif) receptor 4 (CXCR4) N terminus.J Biol Chem. 2013 Jan 4;288(1):737-46. doi: 10.1074/jbc.M112.394064. Epub 2012 Nov 12. J Biol Chem. 2013. PMID: 23148226 Free PMC article.
-
Recent Advances in the Application of Solution NMR Spectroscopy to Multi-Span Integral Membrane Proteins.Prog Nucl Magn Reson Spectrosc. 2009 Nov 1;55(4):335-360. doi: 10.1016/j.pnmrs.2009.07.002. Prog Nucl Magn Reson Spectrosc. 2009. PMID: 20161395 Free PMC article. No abstract available.
-
International Union of Basic and Clinical Pharmacology. [corrected]. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors.Pharmacol Rev. 2013 Nov 11;66(1):1-79. doi: 10.1124/pr.113.007724. Print 2014. Pharmacol Rev. 2013. PMID: 24218476 Free PMC article. Review.
References
-
- Bleul CC, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer TA. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. - PubMed
-
- Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier JL, Arenzana-Seisdedos F, Schwartz O, Heard JM, Clark-Lewis I, Legler DF, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. - PubMed
-
- Murphy PM, Baggiolini M, Charo IF, Hebert CA, Horuk R, Matsushima K, Miller LH, Oppenheim JJ, Power CA. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol Rev. 2000;52:145–176. - PubMed
-
- Loetscher M, Geiser T, O'Reilly T, Zwahlen R, Baggiolini M, Moser B. Cloning of a human seven-transmembrane domain receptor, LESTR, that is highly expressed in leukocytes. J Biol Chem. 1994;269:232–237. - PubMed
-
- Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

