Capturing hammerhead ribozyme structures in action by modulating general base catalysis
- PMID: 18834200
- PMCID: PMC2553840
- DOI: 10.1371/journal.pbio.0060234
Capturing hammerhead ribozyme structures in action by modulating general base catalysis
Abstract
We have obtained precatalytic (enzyme-substrate complex) and postcatalytic (enzyme-product complex) crystal structures of an active full-length hammerhead RNA that cleaves in the crystal. Using the natural satellite tobacco ringspot virus hammerhead RNA sequence, the self-cleavage reaction was modulated by substituting the general base of the ribozyme, G12, with A12, a purine variant with a much lower pKa that does not significantly perturb the ribozyme's atomic structure. The active, but slowly cleaving, ribozyme thus permitted isolation of enzyme-substrate and enzyme-product complexes without modifying the nucleophile or leaving group of the cleavage reaction, nor any other aspect of the substrate. The predissociation enzyme-product complex structure reveals RNA and metal ion interactions potentially relevant to transition-state stabilization that are absent in precatalytic structures.
Conflict of interest statement
Figures
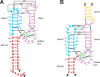


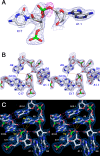
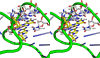
Comment in
-
Slowing down a hammerhead ribozyme allows glimpses of it in action.PLoS Biol. 2008 Sep;6(9):e244. doi: 10.1371/journal.pbio.0060244. Epub 2008 Sep 30. PLoS Biol. 2008. PMID: 20076723 Free PMC article. No abstract available.
References
-
- Nelson JA, Uhlenbeck OC. When to believe what you see. Mol Cell. 2006;23:447–450. - PubMed
-
- Westhof E. A tale in molecular recognition: the hammerhead ribozyme. J Mol Recognit. 2007;20:1–3. - PubMed
-
- Wedekind JE, McKay DB. Crystallographic structures of the hammerhead ribozyme: relationship to ribozyme folding and catalysis. Annu Rev Biophys Biomol Struct. 1998;27:475–502. - PubMed
-
- Blount KF, Uhlenbeck OC. The structure-function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct. 2005;34:415–440. - PubMed

