Mining physical protein-protein interactions from the literature
- PMID: 18834490
- PMCID: PMC2559983
- DOI: 10.1186/gb-2008-9-s2-s12
Mining physical protein-protein interactions from the literature
Abstract
Background: Deciphering physical protein-protein interactions is fundamental to elucidating both the functions of proteins and biological processes. The development of high-throughput experimental technologies such as the yeast two-hybrid screening has produced an explosion in data relating to interactions. Since manual curation is intensive in terms of time and cost, there is an urgent need for text-mining tools to facilitate the extraction of such information. The BioCreative (Critical Assessment of Information Extraction systems in Biology) challenge evaluation provided common standards and shared evaluation criteria to enable comparisons among different approaches.
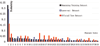
Results: During the benchmark evaluation of BioCreative 2006, all of our results ranked in the top three places. In the task of filtering articles irrelevant to physical protein interactions, our method contributes a precision of 75.07%, a recall of 81.07%, and an AUC (area under the receiver operating characteristic curve) of 0.847. In the task of identifying protein mentions and normalizing mentions to molecule identifiers, our method is competitive among runs submitted, with a precision of 34.83%, a recall of 24.10%, and an F1 score of 28.5%. In extracting protein interaction pairs, our profile-based method was competitive on the SwissProt-only subset (precision = 36.95%, recall = 32.68%, and F1 score = 30.40%) and on the entire dataset (30.96%, 29.35%, and 26.20%, respectively). From the biologist's point of view, however, these findings are far from satisfactory. The error analysis presented in this report provides insight into how performance could be improved: three-quarters of false negatives were due to protein normalization problems (532/698), and about one-quarter were due to problems with correctly extracting interactions for this system.
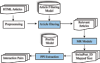
Conclusion: We present a text-mining framework to extract physical protein-protein interactions from the literature. Three key issues are addressed, namely filtering irrelevant articles, identifying protein names and normalizing them to molecule identifiers, and extracting protein-protein interactions. Our system is among the top three performers in the benchmark evaluation of BioCreative 2006. The tool will be helpful for manual interaction curation and can greatly facilitate the process of extracting protein-protein interactions.
Figures



Similar articles
-
Overview of the protein-protein interaction annotation extraction task of BioCreative II.Genome Biol. 2008;9 Suppl 2(Suppl 2):S4. doi: 10.1186/gb-2008-9-s2-s4. Epub 2008 Sep 1. Genome Biol. 2008. PMID: 18834495 Free PMC article.
-
Evaluation of text-mining systems for biology: overview of the Second BioCreative community challenge.Genome Biol. 2008;9 Suppl 2(Suppl 2):S1. doi: 10.1186/gb-2008-9-s2-s1. Epub 2008 Sep 1. Genome Biol. 2008. PMID: 18834487 Free PMC article.
-
Evaluation of BioCreAtIvE assessment of task 2.BMC Bioinformatics. 2005;6 Suppl 1(Suppl 1):S16. doi: 10.1186/1471-2105-6-S1-S16. Epub 2005 May 24. BMC Bioinformatics. 2005. PMID: 15960828 Free PMC article.
-
Analysis of biological processes and diseases using text mining approaches.Methods Mol Biol. 2010;593:341-82. doi: 10.1007/978-1-60327-194-3_16. Methods Mol Biol. 2010. PMID: 19957157 Review.
-
Facts from text: can text mining help to scale-up high-quality manual curation of gene products with ontologies?Brief Bioinform. 2008 Nov;9(6):466-78. doi: 10.1093/bib/bbn043. Epub 2008 Dec 6. Brief Bioinform. 2008. PMID: 19060303 Review.
Cited by
-
Classifying protein-protein interaction articles using word and syntactic features.BMC Bioinformatics. 2011 Oct 3;12 Suppl 8(Suppl 8):S9. doi: 10.1186/1471-2105-12-S8-S9. BMC Bioinformatics. 2011. PMID: 22151252 Free PMC article.
-
A robust data-driven approach for gene ontology annotation.Database (Oxford). 2014 Nov 25;2014:bau113. doi: 10.1093/database/bau113. Print 2014. Database (Oxford). 2014. PMID: 25425037 Free PMC article.
-
Bridging semantics and syntax with graph algorithms-state-of-the-art of extracting biomedical relations.Brief Bioinform. 2017 Jan;18(1):160-178. doi: 10.1093/bib/bbw001. Epub 2016 Feb 5. Brief Bioinform. 2017. PMID: 26851224 Free PMC article.
-
Decoding Plant-Pathogen Interactions: A Comprehensive Exploration of Effector-Plant Transcription Factor Dynamics.Mol Plant Pathol. 2025 Jan;26(1):e70057. doi: 10.1111/mpp.70057. Mol Plant Pathol. 2025. PMID: 39854033 Free PMC article. Review.
-
Overview of the protein-protein interaction annotation extraction task of BioCreative II.Genome Biol. 2008;9 Suppl 2(Suppl 2):S4. doi: 10.1186/gb-2008-9-s2-s4. Epub 2008 Sep 1. Genome Biol. 2008. PMID: 18834495 Free PMC article.
References
-
- Chatr-aryamontri A, Ceol A, Licata L, Cesareni G. Annotating molecular interactions in the MINT database. Proceedings of the BioCreative Workshop; 22 to 25 April 2007; Madrid, Spain. http://compbio.uchsc.edu/Hunter_lab/Cohen/BC2_Proceedings.pdf
-
- Khadake J, Aranda B, Derow C, Huntley R, Kerrien S, Leroy C, Orchard S, Apweiler R, Hermjakob H. IntAct - serving the text-mining community with high quality molecular interaction data. Proceedings of the BioCreative Workshop; 22 to 25 April 2007; Madrid, Spain. http://compbio.uchsc.edu/Hunter_lab/Cohen/BC2_Proceedings.pdf
-
- Hermjakob H, Montecchi-Palazzi L, Lewington C, Mudali S, Kerrien S, Orchard S, Vingron M, Roechert B, Roepstorff AP, Bairoch A, Cesareni G, Sherman D, Apweiler R. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

