Overview of the protein-protein interaction annotation extraction task of BioCreative II
- PMID: 18834495
- PMCID: PMC2559988
- DOI: 10.1186/gb-2008-9-s2-s4
Overview of the protein-protein interaction annotation extraction task of BioCreative II
Abstract
Background: The biomedical literature is the primary information source for manual protein-protein interaction annotations. Text-mining systems have been implemented to extract binary protein interactions from articles, but a comprehensive comparison between the different techniques as well as with manual curation was missing.
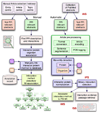
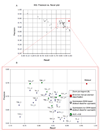
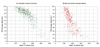
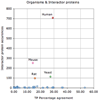
Results: We designed a community challenge, the BioCreative II protein-protein interaction (PPI) task, based on the main steps of a manual protein interaction annotation workflow. It was structured into four distinct subtasks related to: (a) detection of protein interaction-relevant articles; (b) extraction and normalization of protein interaction pairs; (c) retrieval of the interaction detection methods used; and (d) retrieval of actual text passages that provide evidence for protein interactions. A total of 26 teams submitted runs for at least one of the proposed subtasks. In the interaction article detection subtask, the top scoring team reached an F-score of 0.78. In the interaction pair extraction and mapping to SwissProt, a precision of 0.37 (with recall of 0.33) was obtained. For associating articles with an experimental interaction detection method, an F-score of 0.65 was achieved. As for the retrieval of the PPI passages best summarizing a given protein interaction in full-text articles, 19% of the submissions returned by one of the runs corresponded to curator-selected sentences. Curators extracted only the passages that best summarized a given interaction, implying that many of the automatically extracted ones could contain interaction information but did not correspond to the most informative sentences.
Conclusion: The BioCreative II PPI task is the first attempt to compare the performance of text-mining tools specific for each of the basic steps of the PPI extraction pipeline. The challenges identified range from problems in full-text format conversion of articles to difficulties in detecting interactor protein pairs and then linking them to their database records. Some limitations were also encountered when using a single (and possibly incomplete) reference database for protein normalization or when limiting search for interactor proteins to co-occurrence within a single sentence, when a mention might span neighboring sentences. Finally, distinguishing between novel, experimentally verified interactions (annotation relevant) and previously known interactions adds additional complexity to these tasks.
Figures
References
-
- Mishra G, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan T, Menon S, Hanumanthu G, Gupta M, Upendran S, Gupta S, Mahesh M, Jacob B, Mathew P, Chatterjee P, Arun K, Sharma S, Chandrika K, Deshpande N, Palvankar K, Raghavnath R, Krishnakant R, Karathia H, Rekha B, Nayak R, Vishnupriya G, et al. Human protein reference database: 2006 update. Nucleic Acids Res. 2006;34:D411–D414. - PMC - PubMed
-
- Beuming T, Skrabanek L, Niv M, Mukherjee P, Weinstein H. PDZBase: a protein-protein interaction database for PDZ-domains. Bioinformatics. 2005;21:827–828. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

