Accounting for horizontal gene transfers explains conflicting hypotheses regarding the position of aquificales in the phylogeny of Bacteria
- PMID: 18834516
- PMCID: PMC2584045
- DOI: 10.1186/1471-2148-8-272
Accounting for horizontal gene transfers explains conflicting hypotheses regarding the position of aquificales in the phylogeny of Bacteria
Abstract
Background: Despite a large agreement between ribosomal RNA and concatenated protein phylogenies, the phylogenetic tree of the bacterial domain remains uncertain in its deepest nodes. For instance, the position of the hyperthermophilic Aquificales is debated, as their commonly observed position close to Thermotogales may proceed from horizontal gene transfers, long branch attraction or compositional biases, and may not represent vertical descent. Indeed, another view, based on the analysis of rare genomic changes, places Aquificales close to epsilon-Proteobacteria.
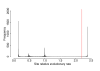
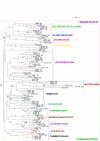
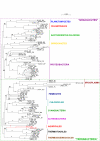
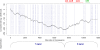
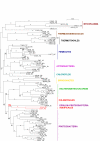
Results: To get a whole genome view of Aquifex relationships, all trees containing sequences from Aquifex in the HOGENOM database were surveyed. This study revealed that Aquifex is most often found as a neighbour to Thermotogales. Moreover, informational genes, which appeared to be less often transferred to the Aquifex lineage than non-informational genes, most often placed Aquificales close to Thermotogales. To ensure these results did not come from long branch attraction or compositional artefacts, a subset of carefully chosen proteins from a wide range of bacterial species was selected for further scrutiny. Among these genes, two phylogenetic hypotheses were found to be significantly more likely than the others: the most likely hypothesis placed Aquificales as a neighbour to Thermotogales, and the second one with epsilon-Proteobacteria. We characterized the genes that supported each of these two hypotheses, and found that differences in rates of evolution or in amino-acid compositions could not explain the presence of two incongruent phylogenetic signals in the alignment. Instead, evidence for a large Horizontal Gene Transfer between Aquificales and epsilon-Proteobacteria was found.
Conclusion: Methods based on concatenated informational proteins and methods based on character cladistics led to different conclusions regarding the position of Aquificales because this lineage has undergone many horizontal gene transfers. However, if a tree of vertical descent can be reconstructed for Bacteria, our results suggest Aquificales should be placed close to Thermotogales.
Figures


References
-
- Wolf YI, Rogozin IB, Grishin NV, Koonin EV. Genome trees and the tree of life. Trends Genet. 2002;18:472–479. - PubMed
-
- Klenk HP, Meier TD, Durovic P, Schwass V, Lottspeich F, Dennis PP, Zillig W. RNA polymerase of Aquifex pyrophilus: implications for the evolution of the bacterial rpoBC operon and extremely thermophilic bacteria. J Mol Evol. 1999;48:528–541. - PubMed
-
- Cavalier-Smith T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int J Syst Evol Microbiol. 2002;52:7–76. - PubMed
-
- Coenye T, Vandamme P. A genomic perspective on the relationship between the Aquificales and the epsilon-Proteobacteria. Syst Appl Microbiol. 2004;27:313–322. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

